
National Institute for Basic Biology





Although transposons occupying a large portion of the genome in various plants were once thought to be junk DNA, they play an important role in genome reorganization and evolution. Active DNA transposons are important tools for gene functional analysis. The endogenous non-autonomous transposon, nDart1, in rice is said to generate various transposon-insertion mutants because nDart1 elements tend to insert into genic regions under natural growth conditions. The transpositions of nDart1 were promoted by an active autonomous element, aDart1-27, on chromosome 6. By using the endogenous nDart1/aDart1-27 system in rice, a large-scale nDart-inserted mutant population was easily generated under normal field conditions, and the resulting tagged lines were free of somaclonal variation. The nDart1/aDart1-27 system was introduced into a rice variety, Koshihikari (Oryza sativa subsp. japonica), and Basmati (Oryza sativa subsp. indica). Various mutations caused by the insertion of nDart have been screened for characteristic phenotypes.
Seed size and number were controlled by various genes in the plants. It was reported that expression changes in high contribution genes for seed size, number and panicle shape resulted in a decrease of the total yield. A strategy for boosting rice yield based on molecular biology is to stack the finely tuned gene expressions. The Lgg mutant which was isolated from Koshihikari-nDart tagging line bore slightly larger grains (Figure 1) as a dominant inheritance. Transposon-display identified the insertion site of nDart1 in the Lgg mutant.

Figure 1. Phenotype of Large grain (Lgg). Harvested panicle and seeds.
The identified LGG gene shows similarity to RNA binding proteins. To investigate the subcellular localization of LGG protein, green fluorescent protein (GFP)-fused constructs driven by 35S promoter, 35S:LGGNP-GFP was transformed into rice calli. GFP fluorescence spots were observed in the nuclei in calli. These results suggest that LGG is localized to the nucleus. To verify that LGG was responsible for the long grain phenotype, knockout (GE) and overexpression (OE) lines were produced by transforming a CRISPR/Cas9 construct targeting the RNA recognition motif region of LGG and an LGG construct with its WT promoter into into cv. Nipponbare (NP), respectively (Figure 2(A)). Several independent GE lines showed significantly increased grain length, and three independent OE lines had shorter grains than NP. Any significant differences of panicle length, number of panicles per plant and number of spikelets per panicle were not observed among NP, GE and OE except for culm length. Observations of longitudinal sections from lemmas of GE and OE lines revealed that the cell sizes of GE and OE lines were comparable to NP (Figure2 B-C), suggesting that LGG might regulate the longitudinal cell number of spikelet hull and thus grain length.
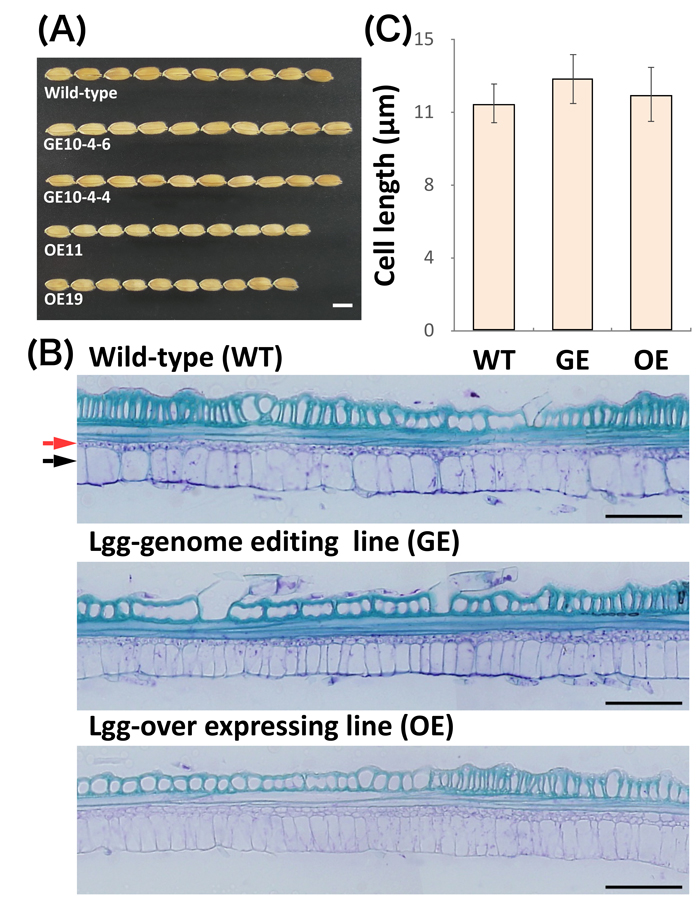
Figure 2. The spikelets morphology and longitudinal sections and cell length of the lemma of NP, GE, and OE, respectively. (A) The spikelets. Bar = 5 mm. (B) The Section of lemma in transgenic plants. The red and black arrows indicate adaxial epidermis and chlorophyll-contained parenchyma cell. (C) Cell length of lemma in NP, GE and OE. n = 3. mean ± SD.