Although DNA transposon is one of major component in plant genomes, its transposition is restricted genetically or epigenetically for genome stability. Because the insertions of transposons have been contributed to the creation of new genes and genome evolution, to reveal the genome dynamisms driven by DNA transposon is purpose of research.
Gene tagging is important tool for understanding gene functions. We have constructed rice mutant line using DNA transposons in order to achieve functional genomics analysis in rice, a model plant for monocots and cereals.
I. An active DNA transposon in rice
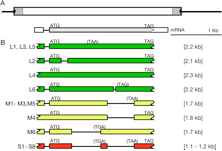
The mutable virescent (or pale-yellow-leaf variegated, pyl-v) which display leaf variegation was caused by the integration of the 607-bp nonautonomous element nDart1-0 belonging to the hAT superfamily into the OsClpP5 genes encoding the chloroplast protease. In the mutable pyl-v allele, somatic excision of nDart1-0 from OsClpP5 in the presence of an active autonomous element, aDart, results in the pyl-v leaf variegation phenotype, a dark-green sector consisting of somatically reverted cells due to nDart1-0 excision in a pale-yellow background comprising cells having nDart1-0 inserted into OsClpP5 in the homozygous condition. Plants containing the pyl-v allele without an active aDart element display pale-yellow leave without variegation; this has been termed as the pale-yellow leaf-stable (pyl-stb) phenotype. In the sequenced Nipponbare genome containing no active aDart elements, among the 53 inactive iDart1 elements, 38 iDart1 elements are putative autonomous element. Because their putative transposase genes carry no apparent nonsense or frameshift mutations, they are thought to be silenced epigenetically. It is, thus, highly likely that an active aDart element is similar in structure to one of these 38 iDart1 elements. By map-based cloning, aDart in the mutable pyl-v plant coincides with one of the 38 iDart1 elements, iDart1-27, residing on chromosome 6 in Nipponbare and that the transcripts of the accumulated transposase gene in the pyl-v leaves are predominantly from Dart1-27. Two additional smaller transcripts could be detected in pyl-v (Figure. 1). The major transcripts detected in pyl-stb or Nipponbare were derived from non-Dart1-27 elements, implying that the residual expression of other Dart1 elements in both pyl-stb and Nipponbare would be too weak to act on the nDart1-0 at OsClpP5 to lead to their excision even though some of them might encode an active transposase. While the longest (L) transcripts in pyl-v contained the Dart1-27 transcripts having intron 1 at 5’-UTR spliced, the middle (M) and the shortest (S) transcripts were mixtures that had one or two additional introns spliced, respectively, although the exact splicing sites were often different. It is noteworthy that all the shorter transcripts characterized in pyl-v were derived from Dart1-27, indicating that some of the abundantly expressed transcripts must have undergone further splicing in rice. These findings would facilitate the development of an efficient gene-tagging system in rice and shed light on epigenetic regulatory and evolutionary aspects of autonomous elements in the nDart/aDart system.
Figure 1 Transcripts of the Dart1 transposase genes detected in rice. (A) Structure of Dart1-27 in the pyl-v genome. (B) Schematic representation of the alternatively spliced transposase transcripts observed in rice plants. Three different transcripts in sizes (L, M, and S) were observed, and their splicing patterns are categorized.
II. Reverse genetic analysis of rice genes
Among active rice DNA transposons, nDart1-0 and its relatives appear to be more suitable than the others for transposon tagging in rice because (1) their transposition can be controlled under natural growth conditions, i.e., the transposition of nDart1-0 can be induced by crossing with a line containing an active aDart element and stabilized by segregating aDart, and (2) nDart1-0 and its relatives are often found at GC-rich regions in the genome and tend to integrate into promoter proximal genic regions. Because it is also obtained dominant and semi-dominant mutants (Figure 2), nDart1-promorted mutant line can contribute to functional genomics in rice.
Figure 2. Semi-dominant bushy dwarf tillers mutant.