
National Institute for Basic Biology





Recent advances in the CRISPR-Cas system now allow for reverse genetics in various organisms. However, it has been hampered by the lack of a simple and efficient method for gene modification in most of the non-model organisms. To overcome this issue, we developed a highly-efficient workflow for gene knockout in the founder using this CIRSPR-Cas. We call the virtually knockout founders “crispants”. Crispant assay provides us with a practical and rapid tool for functional screening of numerous genes of interest beyond the post-genome era (Figure 1).
Despite the practical utility of the knockout technique, there is still room for improvement in the integration of exogenous DNA into a target chromosomal site (i.e. knock-in), which is still somewhat limited in various organisms. Therefore, we are currently developing more efficient and practical knock-in techniques than conventional ones.
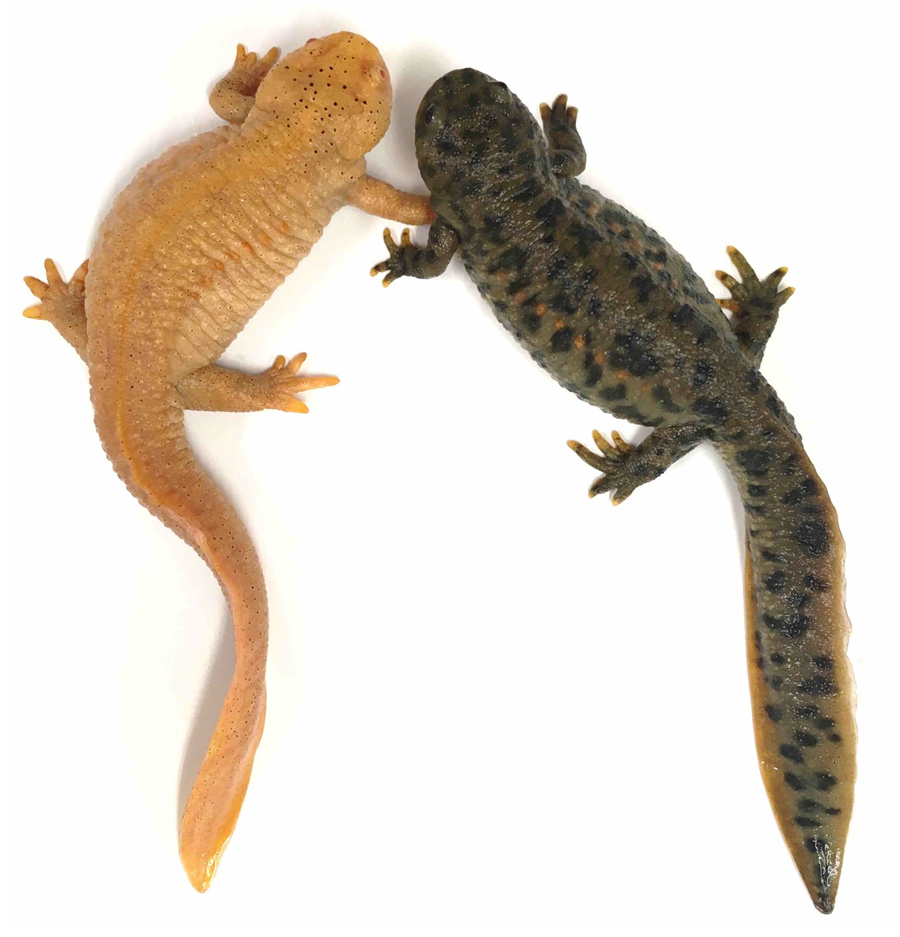
Figure 1. tyrosinase crispant in P. waltl. A knock-out founder of tyrosinase, a melanin synthesis enzyme, and wild newt (left and right, respectively). tyr crispant shows full albinism.
One of our missions is to discover unique organisms and develop them as new model organisms for basic biology. A recent example of this is our recent establishment of the newt Pleurodeles waltl as an experimental model animal for regenerative biology using NGS and genome editing techniques. P. waltl possesses several excellent characteristics as a model animal: easy breeding, short sexual maturation period, remarkable regenerative capacity and comparatively high efficiency of genome editing (Figure 2). We are currently researching the molecular basis of organ regeneration using this newt. In addition, we widely support researchers who attempt to develop new model organisms contributing to the up-coming biology.
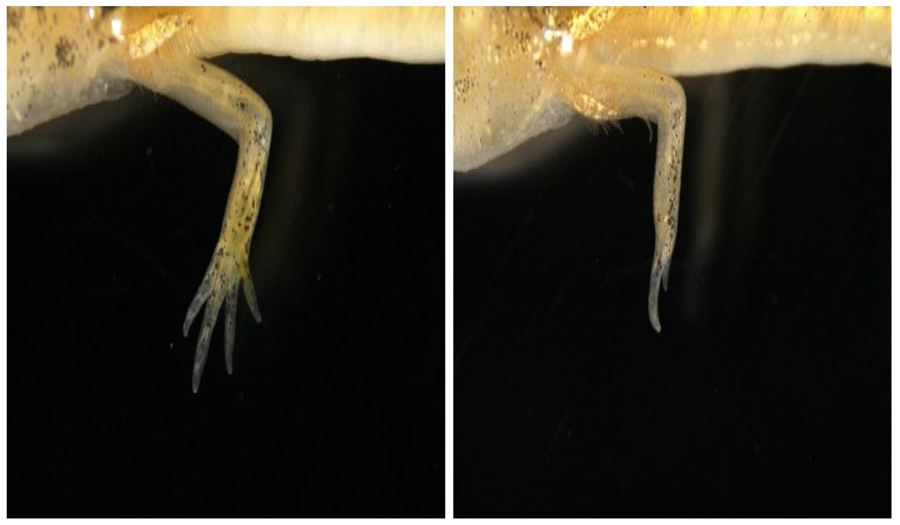
Figure 2. A limb-specific enhancer (ZRS/MFCS1) of sonic hedgehog crispant in P. waltl. Phenotypes of limb regeneration in wild and ZRS/MFCS1 crispant (left and right, respectively). Unlike in normal limb regeneration in the wild type, severe reduction of digit formation was seen in ZRS/MFCS1 crispant.