
National Institute for Basic Biology






A medaka habitat in summer (Miyazaki, Miyazaki Prefecture) (left). A school of wild medaka (Toyohashi, Aichi Prefecture) (right).
The natural environment, which incorporates phenomena such as day length, solar radiation, temperature, and precipitation, generates seasonal changes that affect organisms. Although animals alter their physiology and behavior in response to seasonal changes in their environment, the mechanism of seasonal adaptation remains largely unknown.
Medaka (Oryzias latipes) provide an excellent model to study these mechanisms because of their rapid and obvious seasonal responses. In addition, it is also possible to apply transgenic and genome-editing approaches when researching them, as well as reference genome sequences.
In keeping with this, we are currently using Medaka to study the molecular mechanisms of seasonal adaptation, as well as the relationship between seasonal information from the environment and the organism's response.
Most animals living outside the tropics reproduce only during a particular season of the year. These are called seasonal breeders, and it is well established that day length is a crucial cue for reproduction in many of them. In addition, it has been demonstrated that temperature changes are also important for them in the how they detect seasons. However, it remains unknown how animals measure seasonal changes in relation to these environmental factors.
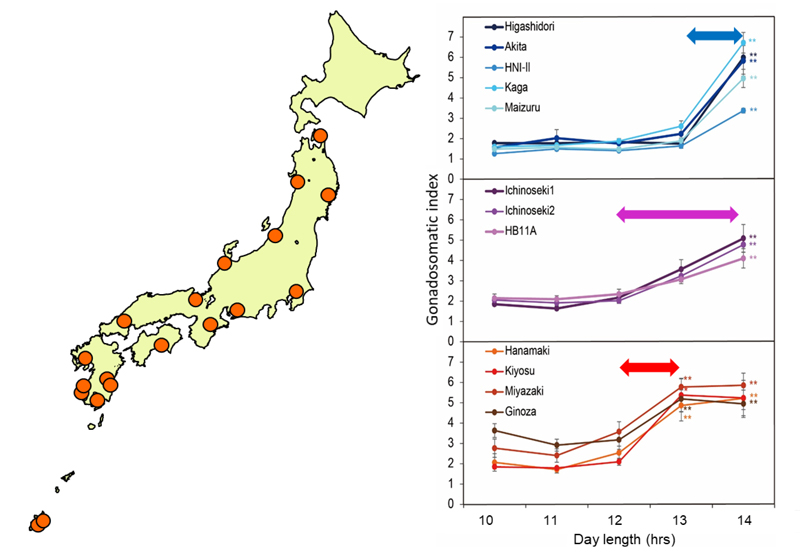
Figure 1. Differences in critical day length (right) were detected among medaka populations that had originated in various latitudes (left) .
Applying forward genetic approaches, we examined critical day length in reproduction (i. e., the duration of lighting time required to cause gonadal development) using various Medaka populations, that originated in various latitudes throughout Japan to elucidate these mechanisms.
Geographical variation critical day length was detected, and populations from lower latitudes indicated a shorter critical day length (Figure 1). To identify the genes governing critical day length, quantitative trait loci (QTL) analysis was conducted using F2 offspring derived from crosses between populations experiencing different critical day length. We thus identified significant QTLs using Restriction-site Associated DNA (RAD) markers (Figure 2).
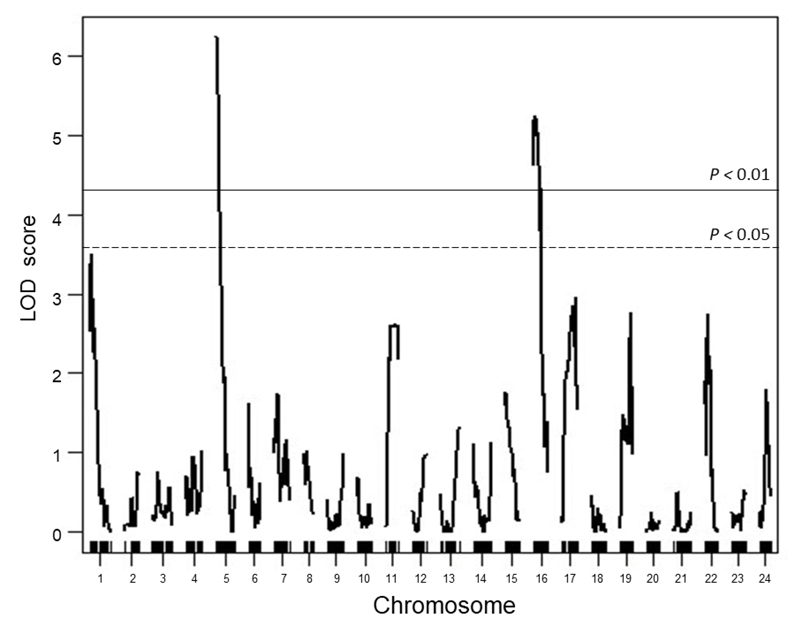
Figure 2. Result of QTL analysis for critical day length.
Whole-genome re-sequencing in various medaka populations experiencing different critical day lengths were conducted. We have identified potential candidate genes that define the critical day length by analyzing this genome sequencing data.
We also performed experiments to identify critical temperature, and subsequently detected geographical variation among Medaka populations. Significant QTLs for critical temperature have been detected from the genetic analysis that was performed.
Organisms exhibit various scales of rhythm, ranging in seconds to years. On the other hand, the natural environment provides the rhythmic changes concerning organisms. However, the quantitative relationship between the information on environmental factors and biological rhythms is poorly understood.
We conducted a linear regression analysis regarding annual rhythms in gonadal development using data pertaining to the annual changes in the gonadal size of the medaka and environmental information (day length, solar radiation, water temperature) in the experimental field. The regression model explained which environmental factors contributed to the seasonal change in medaka gonads and to what extent they contributed to this change.