
National Institute for Basic Biology





Computer programs that accurately capture and measure the features from images that express the phenomenon become an inevitable tool to objectively describe and evaluate a biological phenomenon obtained as digital imaging data. From this point of view, we are developing image analysis programs for each experimental case and analyzing image data that captures various biological phenomena.
To better understand organ formation mechanisms, it is necessary to analyze individual cells’ morphology and dynamics quantitatively. However, it is difficult to achieve it due to the large image volume size generated by such as time-lapse microscopy and its ambiguity.
To unveil organogenesis from the point of view of distinct cell behaviors, we are developing application software capable of describing cell dynamics from time-lapse imaging data sets by employing image processing techniques. To observe this, we are developing a software pipeline which will automatically recognize individual cell shapes out of 3D space and tracks them through time. This system extracts cell boundaries and reconstitutes cell shapes in the form of a skeletonized chain of voxels spanning 3D space. This abstract form of cell visualization makes it possible to describe morphometric quantities and kinetics of cells at a single-cell resolution (Figure 1). These morphometric quantities allow us to perform comparative studies on shapes and behaviors more precisely under several experimental conditions to gain a better understanding of the genetic programs underlying organogenesis. We are now applying this system to several experimental models to determine the practicality of the system.
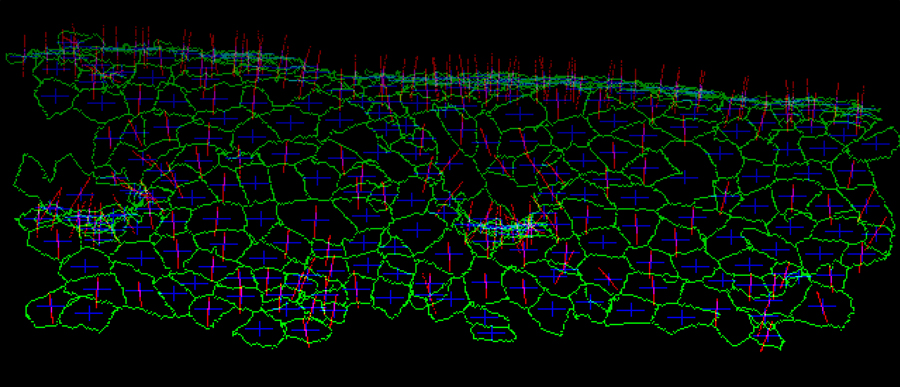
Figure 1. Visualized apical cell surface of Drosophila embryonic epidermal cells. A time-lapse confocal microscopic data set expressing E-cadherin-GFP was processed. Cell boundaries (green), the center of gravity (blue), and normal vector (magenta) are indicated for each cell.
In complex organs, methods for identifying and tracking the initial positions of the cell groups consisting of the final organ morphology are essential analytical methods for elucidating the developmental program.
Here, we analyzed the details of the pathways of individual cells in a brain primordium throughout the brain morphogenesis in chick embryos in its early stages. We identified the cell positions out of randomly fluorescently labeled brain primordium cells from a series of time-lapse images and constructed a database of their time evolution. Based on this database, we developed a software system for automating the visualization and quantification of the local migration pattern of brain primordium constituent cells from several experimental conditions.
III. Quantification of fish body shape
Techniques for quantitatively analyzing the movements and body forms of model organisms are considered indispensable for phenotypic evaluation in the process of elucidating the molecular mechanisms underlying the behavior of living animals.
To elucidate the mechanism of neuronal development of zebrafish, we have developed a system that extracts the body region from each frame of a video of free migration of fish juveniles taken at a high frame rate and describes the morphology from the head to the tail as a free curve model. From these curve models, we developed a system that classifies and simplifies the continuous bending forms of fish as a small number of classes. This system was applied to the movies acquired under various experimental conditions and successfully described and statistical evaluation of the phenotype.