
National Institute for Basic Biology





Tor (target of rapamycin) is a Ser/Thr protein kinase which is well conserved in organisms spanning from yeasts to mammals. Tor forms two distinct protein complexes, TORC1 (Tor complex1) and TORC2. On one hand, TORC1 is involved in amino acid sensing, regulation of protein synthesis (especially the translation step), the cell cycle, and autophagy. On the other hand, TORC2 is responsible for actin organization and cell integrity. Up until now, it’s been deemed unlikely that TORC2 can recognize nutrient signals.
The aim of our research group is to reveal the molecular mechanisms underlying how TORC1 receives amino acid signals and how the TORC1/2 pathways regulate phenomena they are associated with. We have been studying Tor signaling in the budding yeast Saccharomyces cerevisiae, and have found three novel TOR signaling pathway branches (Figure 1).
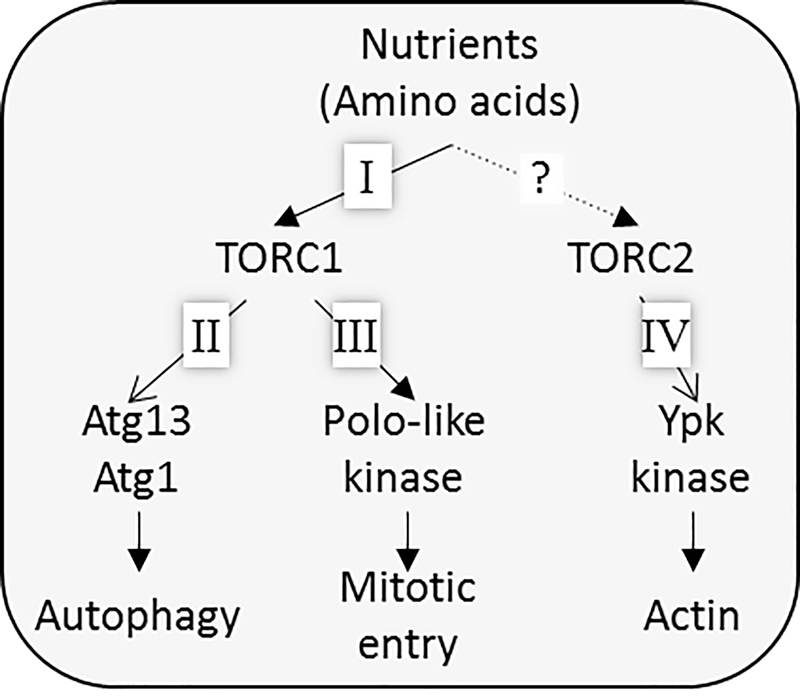
Figure 1. Tor signaling pathway in budding yeast. Our group has found three branches of the Tor pathway.
TORC1 is regulated by amino acids which in themselves are fundamental nutrients. 20 species of amino acids that build proteins cannot be interchanged with each other. Therefore, each amino acid must be individually detected by TORC1.
We have conducted genetic research and discovered the involvement of (aminoacyl-) tRNA in TORC1 regulation. For example, mutants of aminoacyl-tRNA synthetases (ARSs) exhibited inactivation of TORC1 even under amino acid-rich condition, suggesting that aminoacyl-tRNA, a product of ARS acts as an amino acid signal rather than an amino acid itself. Biochemical in vitro TORC1 assay also revealed that tRNA directly inhibits TORC1 kinase activity, suggesting that uncharged tRNA, produced under amino acid-depleted condition, functions as a starvation signal. Reducing cellular tRNA molecules desensitizes TORC1 inactivation by nitrogen starvation in vivo.
Based on these results, a TORC1 regulatory model was proposed which contends that free tRNA released from protein synthesis under amino acid starvation inhibits TORC1 activity. Therefore, TORC1 employs a tRNA- mediated mechanism to monitor intracellular amino acids (Figure 2).
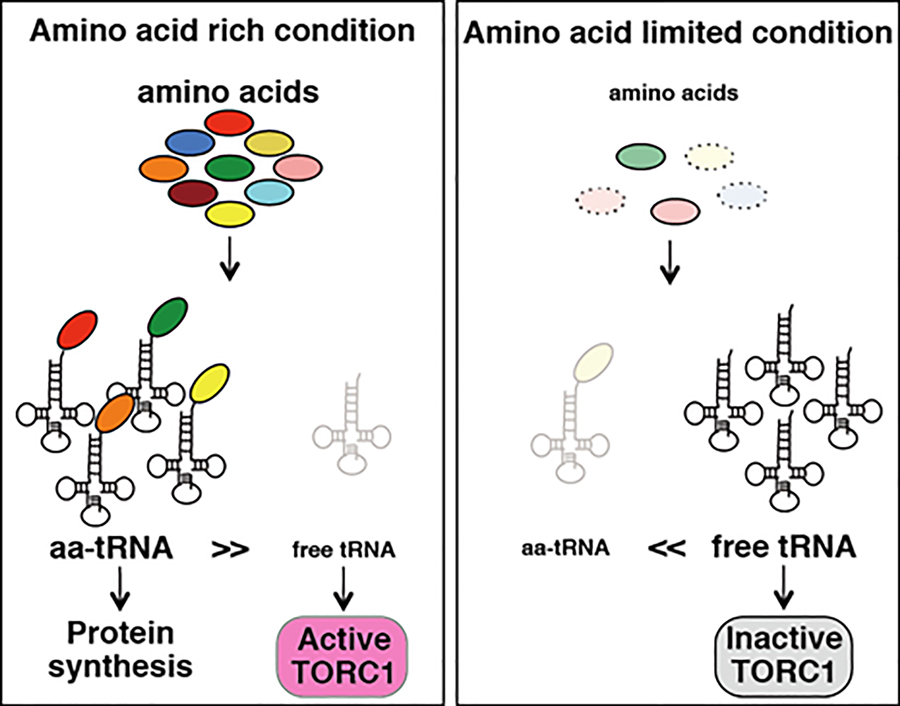
Figure 2. A schematic model of how amino acid is sensed by TORC1. Cytosolic free tRNA inactivates TORC1 under amino acid limited condition.
Since tRNA directly inhibits TORC1 activity, TORC1 should have a tRNA-binding site(s). Thus, we have investigated further so as to determine the tRNA-binding site. So far, we have obtained a good candidate for the tRNA-binding site in Tor protein, and we will now focus on this domain to determine its function.
TORC1 negatively regulates autophagy: a protein degradation system induced by nutrient starvation.
TORC1 negatively regulates autophagy: a protein degradation system induced by nutrient starvation.
We have been consequently able to discover the TORC1-mediated regulatory mechanism of autophagy. Under nutrient-rich conditions, TORC1 directly phosphorylates Atg13, a component of the Atg1 kinase complex. Atg1 is a Ser/Thr protein kinase, the activity of which is essential for autophagy and is largely enhanced in response to TORC1 inactivation. Activation of Atg1 requires formation of the Atg1 complex. Phosphorylation of Atg13 by TORC1 plays a pivotal role in Atg1 complex formation; phosphorylated Atg13 loses its affinity to Atg1 resulting in disassembly of the Atg1 complex and repression of autophagy. On the other hand, dephosphorylation of Atg13 triggers formation of the Atg1 complex, activation of Atg1 kinase, and consequent induction of autophagy.
EIF4E-binding proteins (4EBPs), which binds to translation initiation factor 4E (eIF4E), are thought to negatively regulate translation initiation, because their competition infused binding to eIF4E prevents eIF4E-eIF4G association, which is a primary and essential procedure in translation initiation. In mammals, the inhibitory function of 4EBP1 is regulated by mammalian TORC1 (mTORC1). Under nutrient-rich conditions, mTORC1 phosphorylates 4EBP1, and the phosphorylated 4EBP1, lose affinity to eIF4E and let eIF4E bind to eIF4G.
We examined whether two yeast 4EBPs, Caf20 and Eap1, have properties in common with 4EBP1. Caf20, but not Eap1, is phosphorylated in a TORC1-dependent manner, it binds to eIF4E, and it never associates to eIF4G. However, Caf20 was not directly phosphorylated by TORC1, Caf20-eIF4E binding was not affected by TORC1 activity, and Caf20-eIF4E complex was found in ribosome fraction, suggesting that the function of the yeast 4EBP is different from that of its mammalian counterparts.
Kamada, Y., Umeda, C., Mukai, Y., Ohtsuka, H., Otsubo, Y., Yamashita, A., Kosugi, T. (2024). Structure-based engineering of Tor complexes reveals that two types of yeast TORC1 produce distinct phenotypes. J. Cell Sci. 137, jcs261625.
Kamada, Y., Ando, R., Izawa, S., Matsuura, A. (2023). Yeast Tor complex 1 phosphorylates eIF4E-binding protein, Caf20. Genes to Cells 28, 789-799.
Tai, Y. T., Fukuda, T., Morozumi, Y., Hirai, H., Oda, A. H., Kamada, Y., Akikusa, Y., Kanki, T., Ohta, K., and Shiozaki, K. (2023). Fission yeast TORC1 promotes cell proliferation through Sfp1, a transcription factor involved in ribosome biogenesis. Mol. Cell. Biol. 43, 675-692.
Ando, R., Ishikawa, Y., Kamada, Y., Izawa, S. (2023). Contribution of the yeast bi-chaperone system in the restoration of the RNA helicase Ded1 and translational activity under severe ethanol stress. J. Biol. Chem. 299, 105472.
Kamada, Y. (2017). Novel tRNA function in amino acid sensing of yeast Tor complex1. Genes to Cells 22, 135-147.
Kamada, Y., Yoshino, K., Kondo, C., Kawamata, T., Oshiro, N., Yonezawa, K., and Ohsumi, Y. (2010). Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell Biol. 30, 1049-1058.