I. Condensin-dependent chromatin folding
Chromosome condensation is a basic cellular process that ensures the faithful segregation of chromosomes in both mitosis and meiosis. Any abnormality in this process leads to segregation errors or aneuploidy, which results in cell death. Chromosome condensation is mainly achieved by condensin, a hetero-pentameric protein complex, widely conserved across a variety of organisms ranging from yeast to humans. Despite its conservation and importance in chromosome dynamics, it is not fully understood how condensin works.
Our aim is to understand the mechanism and regulation of chromosome condensation. To this end, we have been studying the role of condensin in the budding yeast
Saccharomyces cerevisiae. We have found that condensin specifically binds to the RFB site located within the rDNA repeat. To date, the best characterized condensin binding region is the rDNA repeat on the right arm of chromosome XII in budding yeast. We further discovered the multiple protein network required to recruit condensin to the RFB site.
The RFB site, which consists of a ~150bp DNA sequence, functions as a cis-element for the recruitment of condensin to chromatin in the yeast genome. If the RFB site is inserted into an ectopic chromosomal locus, condensin can associate with the ectopic RFB site. To explore the role of condensin in chromosome organization, we have constructed a strain in which two RFB sites are inserted into an ectopic chromosome arm with an interval of 15kb distance in the cell with a complete deletion of chromosomal rDNA repeat. Using this strain, condensin-dependent chromatin interaction between two RFBs was examined by chromosome conformation capture (3C) assay. We discovered the condensin-dependent chromatin interaction between the two RFB sites on the chromosome arm. This result indicates that condensin plays a role in chromatin interaction between condensin binding sites, and this interaction leads to the creation of a chromatin loop between those sites (Figure 1). It is thought that condensin-dependent chromatin folding is one of the basic molecular processes of chromosome condensation. During the cell cycle stages, the RFB - RFB interaction signal increases in metaphase and reaches its maximum level in anaphase. In addition to the RFB - RFB interaction, the chromatin interactions between the internal regions of the two RFBs increases in anaphase. Thus, the configuration of chromatin fiber changes from a simple loop into a complicated twisted shape as the cell cycle progresses from metaphase to anaphase.
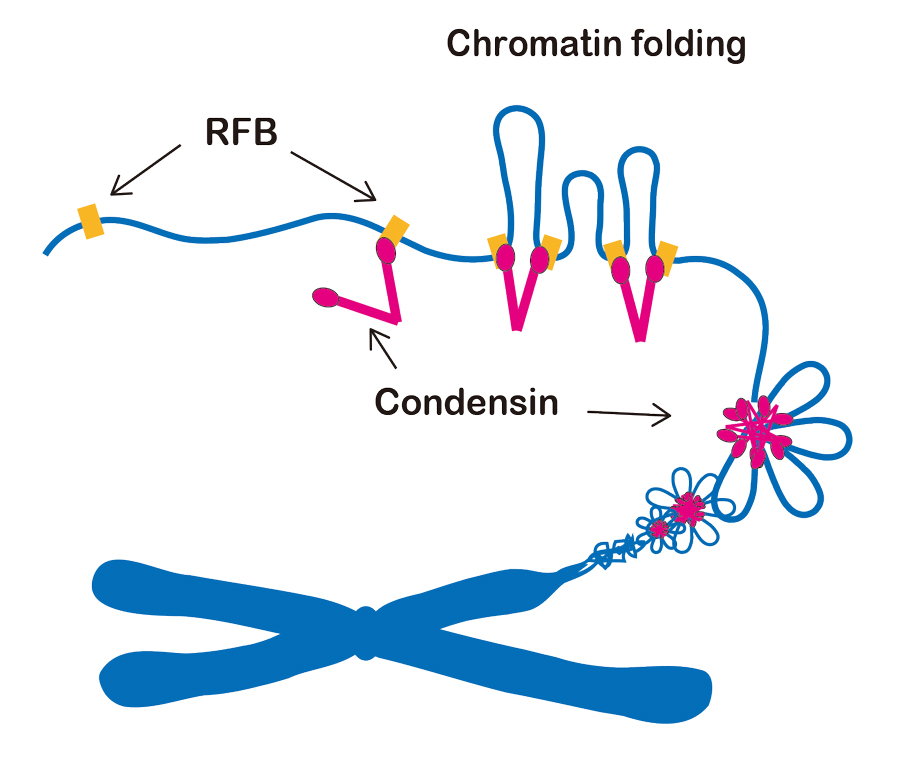
Figure 1. A Schematic model of chromosome condensation. Condensin makes chromatin interactions between adjacent binding sites (RFB, for example). This leads to a folding of chromatin fibers between the sites, as a basic process of chromosome condensation.
II. Plasma-induced cellular responses
Plasma is an ionized gas, which consist of ions and electrons and is extremely reactive. Our question is to understand the effect of the plasma on living organisms.
We have developed a new plasma irradiation device that can control the temperature to the optimal growth temperature of cells, and have developed a system to study the effects of direct plasma exposure to cells without heat stress (Figure 2). Using yeast,
S.cerevisiae, it was found that direct plasma exposure treatment increases lethality as the processing time increases (Figure 3). Furthermore, a plasma-resistant mutant was isolated and the causative gene responsible for plasma-resistance was identified. We are studying functional analysis of the gene to understand the molecular mechanisms of the cellular responses to plasma.
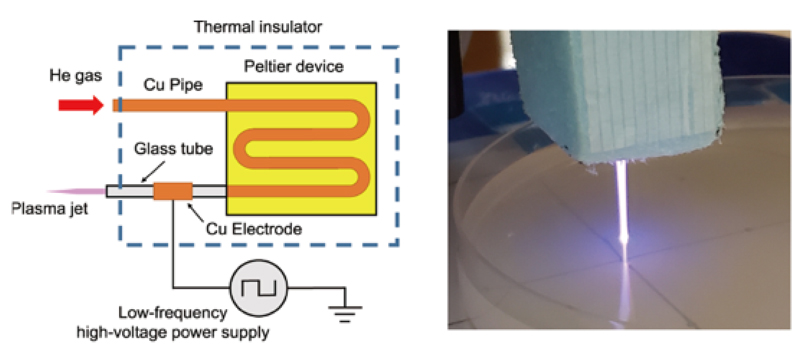
Figure 2. Schematic diagram of atomosperic pressure plasma irradiation device (left). Direct plasma irradiation experiment (right).
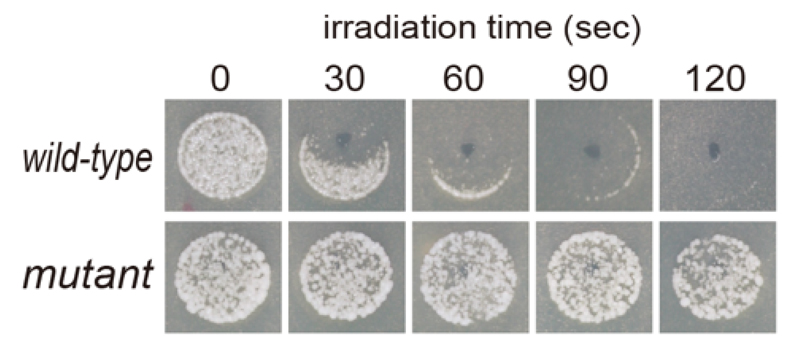
Figure 3. wild-type (upper panels) and plasma-resistant mutant (lower panels)








