Genomic structures and their genetic information are stably transmitted into daughter cells and future generations during cell division; however, they can vary genetically and/or epigenetically. Such variability impacts gene expression and evolution. To understand these genomic dynamics in eukaryotes, particularly in plants, we are analyzing the flower pigmentation of morning glories, including Ipomoea nil (Japanese morning glory), I. purpurea (the common morning glory) and I. tricolor.
I. Flower pigmentation patterns
The wild type morning glories produce flowers with a uniformly pigmented corolla. For our study, we collected mutants displaying pigmentation patterns. Flower pigmentation patterns are easily observable and the molecular mechanisms underlying these phenomena provide useful model systems for investigating genomic variability.
Recessive mutations, duskish of I. nil and pearly-v of I. tricolor, confer variegated flowers caused by a stable insertion of a transposable element into a gene for flower pigmentation, regulated by epigenetic mechanisms (Figure 1). We intend to analyze detailed molecular mechanisms of these mutations.

Figure 1. The
duskish mutant of
I. nil shows variable flower phenotypes and produces variegated, fully pigmented, and pale grayish-purple flowers. It segregates offspring that only show fully pigmented or pale grayish-purple flowers, which can be stably inherited by further generations.
II. Whole-genome analysis of various I. nil lines
The National BioResource Project (NBRP) Morning Glory (described below) maintains ~3,500 lines, including 2 standard lines: the Tokyo Kokei Standard and Violet. A high-quality draft whole-genome sequence of the Tokyo Kokei Standard, which is accessible from our database, has been published. The NBRP Genome Information Upgrading Program supported whole-genome sequencing of 100 representative lines. We chose lines based on the needs of the research community and to provide information regarding their polymorphisms and gene mutations. We included the multiple mutants called “Henka-Asagao,” which covered the major mutations, and the wildtype plants isolated from natural populations outside Japan. From this, we discovered a total of 25,000 Tpn1 transposon-induced insertion polymorphisms and SNPs, Indels, and CNVs at 25 million loci. Several of those polymorphisms were tightly linked to morphological mutations, including the mutations that “Henka-Asagao” carry.
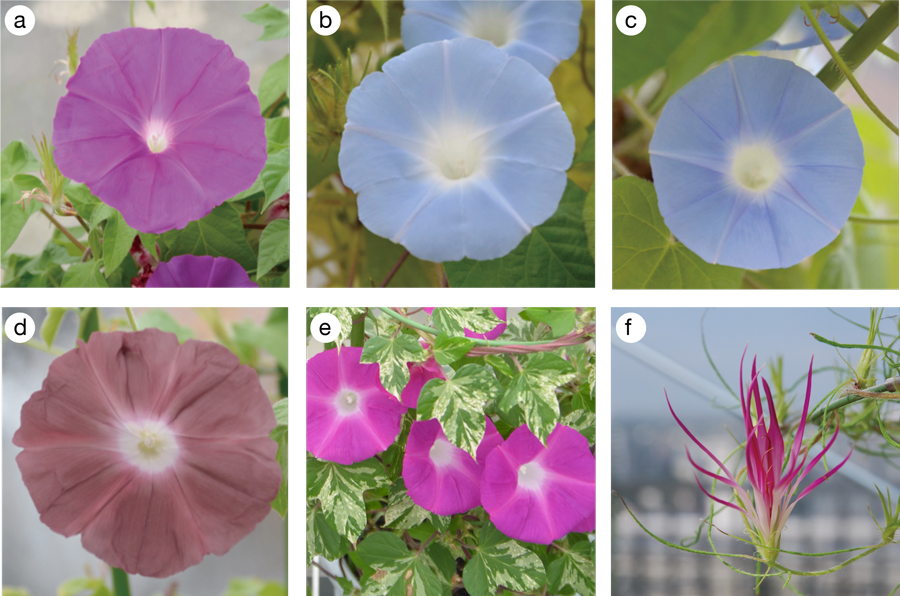
Figure 2. The
I. nil lines used for the whole-genome analysis. (a) Violet: The most widely used standard line. (b) Africa: The wildtype line isolated in Guinea in 1956. (c) Nepal: The wildtype line isolated by Dr. Sasuke Nakao in Nepal in 1952. (d) Danjuro, one of the most popular cultivars, named after the reddish-brown color of the costume used by Danjuro Ichikawa II in his Kabuki play “Shibaraku.” (e) The
tiny mutant has small organs. White spots on the leaves are due to the
variegated mutation. (f) A mutant line of “Henka-Asagao” showing double flowers with narrow flower petals and sepals.
III. Flower opening and time control
The time of flower opening is important for the reproductive strategy of plants, and many plants are known to flower at specific times during the day. However, little is known about the molecular mechanisms that determine flower opening time as well as flower opening itself. As their name suggests, morning glories typically bloom in the early morning. There is significant physiological knowledge about the circadian rhythm that controls flower opening time. To elucidate these molecular mechanisms, we have begun a functional analysis of genes for petal opening and the circadian rhythm.
IV. Morning glory bio-resources
NIBB is the subcenter for the NBRP dedicated to morning glories. In this project, we collect, maintain, and distribute standard and mutant lines for flower pigmentation and DNA clones from EST and BAC libraries of
I. nil and its related species.
I. nil is one of the most popular floricultural plants in Japan and has a 100-year history of extensive related genetic studies. Our collection includes 241 lines and 177,000 DNA clones. The whole-genome sequence, the transcriptome sequences, and the end sequences of the DNA clones can be viewed via the
I. nil genome database (
http://viewer.shigen.info/asagao/index.php).








