The accumulation of biological data has recently been accelerated by various high-throughput “omics” technologies including genomics, transcriptomics, and proteomics. The field of genome informatics is aimed at utilizing this data, or finding the principles behind it, in order to understand complex living systems by integrating the data with current biological knowledge via the use of various computational techniques. In this laboratory we focus on developing computational methods and tools for comparative genome analysis, which is a useful approach for finding functional or evolutionary clues to interpret the genomic information of various species.
The current focus of our research is the comparative analysis of microbial genomes, the number of which has been increasing rapidly. Since different types of information about biological functions and evolutionary processes can be extracted by comparing genomes at different evolutionary distances, we are developing methods to conduct comparative analyses not only of distantly related but also closely related genomes.
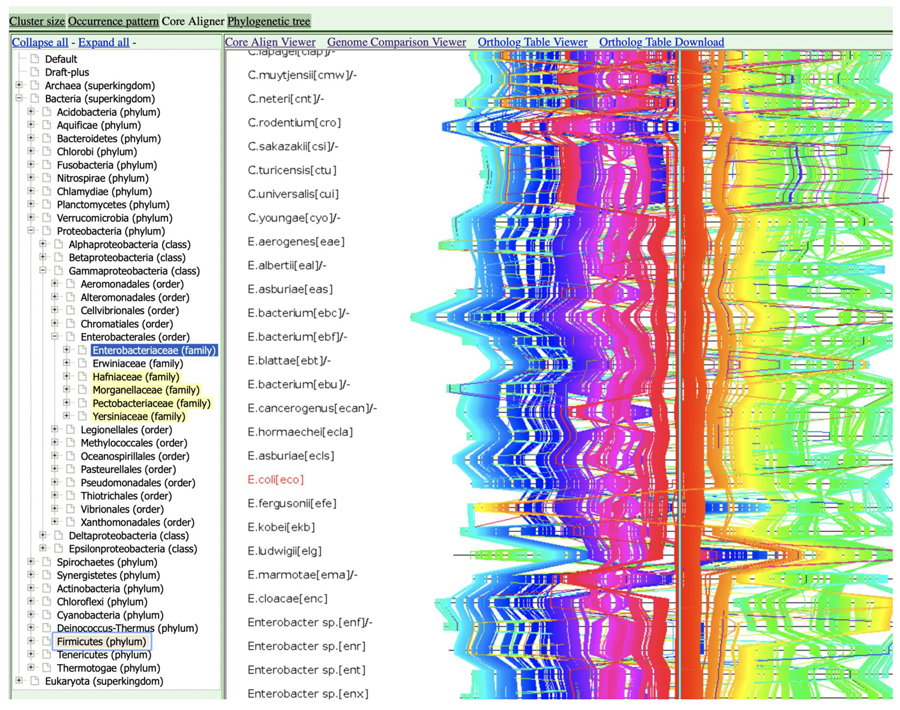
Comparative genome map among the family Enterobacteriaceae on the basis of the core genome alignment constructed using a novel implementation of the CoreAligner program. The color is assigned according to the gene order in Escherichia coli.
Research Projects
-
• Development of the microbial genome database for comparative analysis.
-
• Ortholog classification methods at the domain level and their application.
-
• Comparative analysis of core genome and non-core genome structures among distantly related microbes.
-
• Functional module identification based on the phylogenetic profile method.
-
• Comparative transcriptome analysis between different species.
I. Microbial genome database for comparative analysis
The microbial genome database for comparative analysis (MBGD,
https://mbgd.genome.ad.jp/) is a comprehensive platform for microbial comparative genomics. The central function of MBGD is to create orthologous groups among multiple genomes using the DomClust program combined with the DomRefine program. Through the application of these programs, MBGD not only provides comprehensive orthologous groups among the latest genomic data, but also allows users to create their own ortholog tables on the fly by using a specified set of organisms. MBGD also has pre-calculated ortholog tables for each major taxonomic group, and provides several viewing modes to display the entirety of each ortholog table. For closely related taxa, MBGD provides conserved synteny information calculated using the CoreAligner program. MBGD additionally provides a ‘MyMBGD’ mode, which allows users to add their own genomes to MBGD.
We continue to update the database and MBGD now contains 15397 genomes, including 14786 bacteria, 336 archaea, and 275 eukaryota. These data sets are classified based on the hierarchical ortholog classification strategy described in the section below. In addition, in the latest version of MBGD, we improved several functionalities including a new display of the core genome alignment (Figure 1) and a novel interface for the MyMBGD mode.
As an application software for analyzing novel genome sequences based on the database, we have developed a tool to predict the functional potential of novel microbial genomes through orthology assignment based on the MBGD ortholog table. To evaluate the metabolic potential of the query genome from this assignment, we utilized the Genomaple software to calculate the module completion ratio for each KEGG Module entry (in collaboration with Dr. Takami, Univ Tokyo). The result is displayed on a module completion table where a user can compare the presence or absence of functional modules among specified organisms.

Figure 1. Comparative genome map among the family
Enterobacteriaceae on the basis of the core genome alignment constructed using a novel implementation of the CoreAligner program. The color is assigned according to the gene order in
Escherichia coli.
II. Hierarchical strategy for creating ortholog tables
MBGD previously calculated all-against-all similarities among the stored genomes and independently created two types of ortholog tables: the standard ortholog table containing one representative genome from each genus covering the entire taxonomic range, and the taxon specific ortholog tables containing the genomes belonging to each taxonomic group (species, genus, family and so on).
To create more comprehensive ortholog classification, we developed a stepwise protocol to construct orthologous relationships. First, for each species with at least two genomes, all-against-all similarities among the genomes belonging to that species are calculated and a within-species ortholog table is created. The species-level pan-genome is then created by picking one representative gene from each orthologous group. Next, for each genus with at least two species, all-against-all similarities among the species-level pan-genomes are calculated and a within-genus ortholog table is created. The genus-level pan-genome is then created by picking one representative gene from each orthologous group. Finally, all-against-all similarities among the genus-level pan-genomes are calculated and the standard ortholog table covering the entire taxonomic range is created.
III. Orthologous gene classification among multiple genomes at the domain level
As a core technology of our comparative genomics tools, we have developed a rapid automated method of ortholog grouping, named DomClust, which allows us to simultaneously compare numerous genomes. This method classifies genes based on a hierarchical clustering algorithm using all-against-all similarity data as an input, but it also detects domain fusion or fission events and splits clusters into domains when required. As a result, the procedure can split genes into the domains minimally required for ortholog grouping.
We have also developed a procedure to refine the DomClust classification based on multiple sequence alignments instead of pairwise sequence alignments. The method is based on a scoring scheme, the domain-specific sum-of-pairs (DSP) score, which evaluates domain-level classification using the sum total of domain-level alignment scores. On the basis of this idea, we have developed a refinement pipeline to improve domain-level clustering, DomRefine, by optimizing DSP scores. DomRefine is used to construct the standard ortholog table covering all the representative genomes stored in MBGD.
Domain-level classification is a unique feature within our ortholog classification system. In particular, this data is considered suitable for analyzing domain fusion events that have occurred during evolution. By using the domain-level ortholog grouping data combined with taxonomic and functional information, we are trying to elucidate when and in what kind of genes domain fusion events frequently occur during evolution, and how the complexity of the “domain-fusion network” can be associated with the phenotypic traits in each organism.
IV. Development and application of a workbench for comparative genomics and transcriptomics
We have developed a comparative genomics workbench named RECOG (Research Environment for COmparative Genomics), which aims to extend the current MBGD system by incorporating more advanced analysis functionalities. The central function of RECOG is to display and manipulate large-scale ortholog tables. The ortholog table viewer is a spreadsheet like viewer that can display an entire ortholog table containing more than a thousand genomes. In RECOG, several basic operations on the ortholog table are defined, which are classified into categories such as filtering, sorting, and coloring. By combining these basic operations, various comparative analyses can be performed. In addition, RECOG allows the user to input arbitrary gene properties and compare these properties among orthologs in various genomes.
We are continuing to develop the system and apply it to various genome comparison studies as part of various collaborative research projects. Among these, we applied RECOG to the comparative analyses of transcriptomic data of
Chattonella antiqua and other harmful algae causing red tide in collaboration with Dr. Shikata (FRA). In this analysis, we compared five RNA-seq datasets of harmful algae and 10 existing genome sequences of various algae and a plant model organism,
Arabidopsis thaliana using RECOG, and identified common sequence features among orthologous genes belonging to harmful algae that may be related to red tide outbreaks and their toxicity to fish. The resulting data is available through the database DB-HABs (
https://hab.nibb.ac.jp).
V. Ortholog data representation using the Semantic Web technology to integrate various microbial databases
Orthology is a key to integrating knowledge of various organisms through comparative analysis. In order to integrate genomic data and various types of biological information with this idea, we have constructed an ortholog database using Semantic Web technology. To formalize the structure of the ortholog information in the Semantic Web, we developed the Orthology Ontology (ORTH) and described the ortholog information in MBGD in the form of the Resource Description Framework (RDF).
On the basis of this framework, we have integrated various kinds of microbial data using the ortholog information as a hub, as part of the MicrobeDB.jp project (
http://microbedb.jp/) under the auspices of the National Bioscience Database Center.
VI. A novel approach for identification of genomic islands
Genomes of bacterial species can show great variation in their gene content, and thus systematic analysis of the entire gene repertoire, termed the “pan-genome”, is important for understanding bacterial intra-species diversity. As we have already developed a procedure (CoreAligner) to define the core genome as the genes conserved among the genomes of the given species, we are now focusing on the remaining part of the genomes (non-core genomes) that is more directly linked to the within-species genome diversity. We are developing a method (FindIsland) to identify a set of non-core genes that have conserved gene order by using a modified version of the CoreAligner algorithm. We applied the method to the sets of genomes of prokaryotic species stored in MBGD and developed a database for analyzing their non-core genomes. Based on the database, we found that the resulting conserved clusters frequently correspond to known mobile genetic elements and/or have sequence features common to known genomic islands.







