Pluripotent stem cells (PSCs) are capable of producing all cell types that compose our bodies. In vivo, PSCs appear only transiently during the early stages of development and are lost as development proceeds. Molecular features underlying pluripotency are being extensively studied, and accumulating evidence suggests that the unique regulation of the cell cycle in PSCs is closely linked to the maintenance of pluripotency. We have been investigating DNA replication mechanisms in mammalian PSCs and found that they operate more replication complexes along the genome, although each replication complex synthesizes DNA at a slow rate. This feature is important for mouse embryonic stem (ES) cells, PSCs derived from an early embryo, to coordinate the completion of DNA replication with cell cycle progression. We are currently exploring the molecular link between pluripotency and the unique DNA replication system.
In parallel, our lab investigates how lineage-committed cells can de-differentiate and acquire pluripotency, a process known as “reprogramming.” We utilize a cell-fusion system that allows for the initiation of reprogramming within 24 hours and have established multiple systems to study fused cells at the single-cell level. Our future work aims to provide important insights into the mechanisms of lineage commitment, pluripotency, and reprogramming, as well as to understand the genome instability that is arise during the establishment of induced pluripotent stem (iPS) cells.
Research Projects
-
• Molecular basis of DNA replication mechanisms unique to pluripotent stem cells and their link to nucleotide metabolism
-
• Genome-wide analysis of replication-initiation sites in mouse ES cells
-
• Single-cell profiling of reprogramming cells induced by cell fusion
-
• Cellular behavior of multi-nuclear cells induced by cell fusion
I. Self-renewal of Embryonic Stem Cells and Their Genome-Maintenance Mechanisms
Embryonic stem (ES) cells are derived from the blastocyst stage of embryonic development, and are capable of differentiating into all of the cell types that compose our body (
i.e., ES cells are “pluripotent”). Recent reports suggest that, compared to other cell types, DNA replication machinery of ES cells proceed at lower speed, a feature generally infer presence of replication obstacles. However, the direct cause and the underlying mechanism remains to be uncovered. To date, studies on cell cycle regulation in ES cells have not been as straightforward compared to that of other cell types, as many commonly used cell-synchronization protocols are ineffective for ES cells. We have now established several protocols to synchronize ES cells (Tsubouchi
et al., Cell, 2013; unpublished), which has allowed us to investigate specific stages of the ES cell cycle. So far, we have found that DNA replication is regulated differently in ES cells, to the extent that DNA replication of the whole genome is more accurate in ES cells. We are currently aiming to address how such differences are interlinked with pluripotency by carrying out side-by-side analyses between ES cells and differentiated cells.
II. Induction of Pluripotency through Nuclear Reprogramming and Role of Cell Cycle Regulation
In mammals, PSCs are not maintained in a fully-developed organism. However, differentiated cells can regain pluripotency upon experimental trigger, albeit at a low efficiency. Factors that limit active reprogramming, and conditions that potentiate reprogramming, are the subjects under active investigation.
One of the main obstacles when investigating molecular mechanism underlying reprogramming is the time it takes to start seeing any sign of reprogramming. In order to overcome this problem, we are taking advantage of the cell-to-cell fusion approach, in which a target cell is fused to a pluripotent stem cell to induce pluripotency within a target nucleus, in a short time frame. The cell fusion system is a simple, versatile way to induce reprogramming towards another lineage, and is not limited to pluripotency. Using this system, the first sign of reprogramming can be detected within one day after fusion, thus allowing us to monitor the initial events of reprogramming after induction.
Using this system, we previously found that DNA synthesis is an important event for successful reprogramming (Tsubouchi
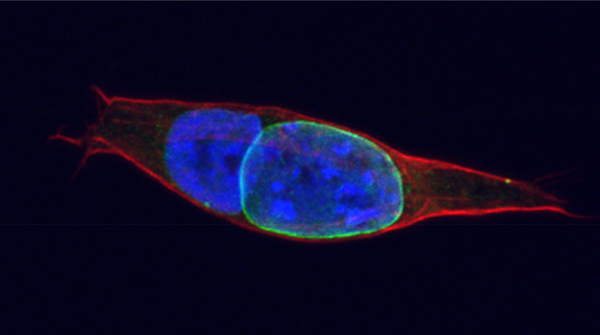
et al., Cell, 2013). Recent reports indicate that reprogramming may cause genetic instability, some of which are thought to arise as DNA replication errors. To investigate the nature of such errors and how they are linked to reprogramming-specific events, we have set up a system to isolate and track a single fused cell (Figure 1) through live-imaging.
Figure 1. Cellular fusion to study reprogramming: a human lymphoblastoid nucleus can be induced to undergo nuclear reprogramming towards pluripotency upon fusion with mouse ES cells (green). Lamin B1 is endogenously tagged with GFP in ES cells, allowing us to distinguish ES vs lymphoblastoid nucleus (unpublished).
III. Future Perspectives
While the fundamental mechanisms that maintain genome integrity have been widely studied using various models, the danger a cell might face when its identity is being altered (through differentiation, reprogramming etc.) are largely unknown. Recent studies of cancer genome sequencing repeatedly identified mutations in the factors that govern cellular identities, leading us to hypothesize that cells may experience genome instability when their identity is unstable. Our goal is to gain a comprehensive understanding of how genome integrity is maintained in ES cells and cells undergoing reprogramming.