“Nothing, it seems, exists except as part of a network of interactions.” (Gilbert & Epel, 2008)
Every creature on Earth exists among a network of various biological interactions. For example, many multicellular organisms, including humans, harbor symbiotic bacteria in their bodies. Some of them provide their hosts with essential nutrients deficient in the host’s diet and others digest foods that are indigestible by the host alone. Despite numerous examples of symbioses and their intriguing outcomes, the genetic and molecular basis underlying these interactions remains elusive. The goal of our group is to establish a new interdisciplinary science known as “Symbiogenomics”, where we aim to understand the network of biological interactions at the molecular and genetic levels. To this end, we take advantage of state-of-the-art genomics, such as next-generation sequencing technologies and CRISPR-Cas9 genome editing.
Research Projects
-
• Molecular mechanisms underlying aphid / Buchnera endosymbiosis
-
• Genome evolution of symbiotic bacteria
-
• Development of genome editing techniques for non-model insects
-
• Application of multi-omics to elucidate symbiotic systems
-
• Genomic basis of sociality evolution in eusocial insects
-
• Bioinformatics and deep-learning approach to understand genetic architecture
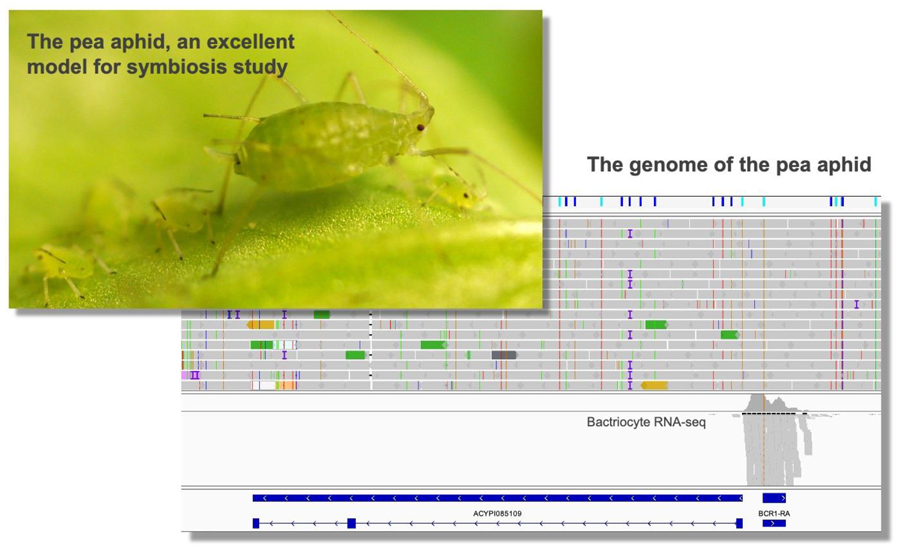
Genomic revelations of a mutualism: the pea aphid and its obligate bacterial symbiont
Aphid species bear intracellular symbiotic bacteria in the cytoplasm of bacteriocytes, which are specialized cells for harboring said bacteria. This mutualism is so obligate that neither can reproduce independently. The genome sequence of the pea aphid,
Acyrthosiphon pisum, in consort with that of bacterial symbiont
Buchnera aphidicola illustrates the remarkable interdependency between these two organisms (IAGC, 2010; Shigenobu
et al., 2000; Shigenobu & Yorimoto, 2022). The genetic capacities of the pea aphid and the symbiont for amino acid biosynthesis are complementary. Genome analysis revealed that the pea aphid has undergone characteristic gene losses and duplications. The IMB antibacterial immune pathway is missing several critical genes, which might account for the evolutionary success of aphids in obtaining beneficial symbionts. Lineage-specific gene duplications have occurred in genes over a broad range of functional categories, which include signaling pathways, miRNA machinery, chromatin modification and mitosis. The importance of these duplications for symbiosis remains to be determined. We found several instances of lateral gene transfer from bacteria to the pea aphid genome. Some of them are highly expressed in bacteriocytes.
We recently discovered a novel class of genes in the pea aphid genome that encode small cysteine-rich proteins with secretion signals that are expressed exclusively in the bacteriocytes of the pea aphid, and named these bacteriocyte-specific cysteine-rich proteins (BCR) (Shigenobu & Stern, 2013). The BCR mRNAs are first expressed at a developmental time point coinciding with the incorporation of symbionts strictly in the cells that contribute to the bacteriocyte, and this bacteriocyte-specific expression is maintained throughout the aphid's life. Furthermore, some BCRs showed antibiotic activity (Uchi
et al., 2019). These results suggest that BCRs act within bacteriocytes to mediate the symbiosis with bacterial symbionts, which is reminiscent of the cysteine-rich secreted proteins of leguminous plants that also regulate endosymbionts. Employment of small cysteine-rich peptides may be a common tactic of host eukaryotes to manipulate bacterial symbionts.
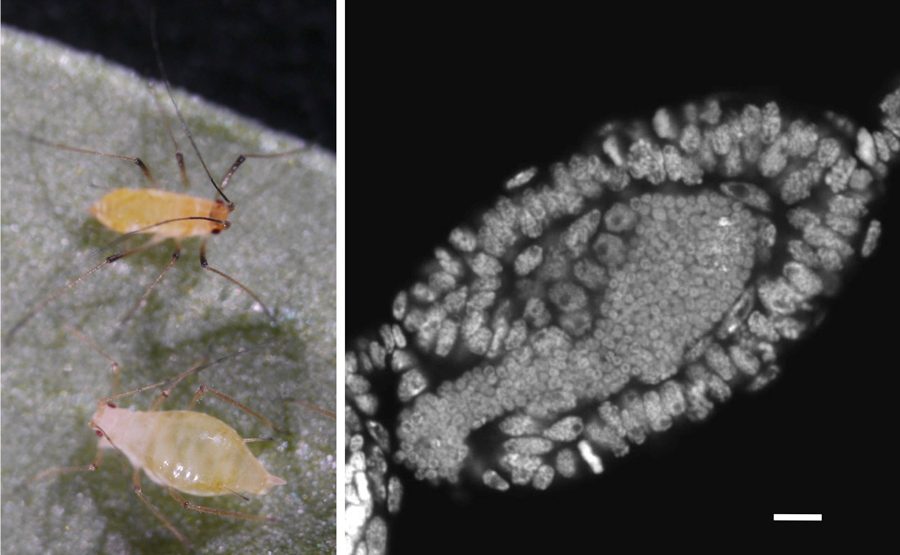
Figure 1. Pea aphids and the bacterial symbiont,
Buchnera. Adult aphids (Left). A developing viviparous embryo which symbionts are infecting (Right). Scale bar = 20 um.
Genomics of emerging model insects
Recent remarkable technological advancements allow us to study any organisms at molecular and genetic levels, which has been difficult for non-model organisms. Insect science is also benefited by the technological innovation. New entomology is starting to study the amazing variety of morphology, phisiology and ethology of insects by using these state-of-the-art technologies. Our group is developing a platform for emerging model insects to read the genomes/transcriptomes and then manipulate their genomes.
Termites are emerging model social organisms characterized by a sophisticated caste system. We recently sequenced the genome of the subterranean termite
Reticulitermes sepratus (Shigenobu et al., 2022). The analyses of ~1.0 Gb genome and 15,591 genes revealed the significance of gene duplication in social evolution. Gene duplication associated with caste-biased gene expression was prevalent in the termite genome. The duplicate genes comprised diverse categories related to social functions, such as chemical communication, social immunity and defense, and they were often expressed in a caste-specific organ. Gene duplication may facilitates social evolution through regulatory diversification leading to caste-biased expression and functional specialization.
Beyond insect genomics, we are involved in several genome projects of emerging model organisms. We recently published the genome information of a photosynthetic sea slug. Some sea slugs take up chloroplasts from the algae that they consume into their cells. These chloroplasts retain their ability to perform photosynthetic activity within the animal cells for several months, and thus provide them with photosynthesis-derived nutrition. This process is called kleptoplasty. Our genome analysis of
Plakobranchus ocellatus revealed the chloroplast acquisition without gene transfers in the photosynthetic sea slug (Maeda et al., 2021).
Caste development and symbiosis in the social aphid Ceratovacuna japonica
Eusociality has evolved repeatedly among insect lineages and represents one of the major transitions in evolution. Molecular mechanisms underlying sociality have been extensively studied with hymenopteran lineages, such as ants, bees, and wasps, and termites. Some aphid species show eusociality and important groups for better understanding of the eusocial evolution, since they are one of the primitive eusocial species that belong to Hemiptera. We focus on the social aphid
Ceratovacuna japonica, because we recently established the laboratory culture system. First, we investigated the sterility regulation in the soldier caste of the aphids (Chung and Shigenobu, 2022). We found that soldiers, that are completely sterile, possess a pair of ovarioles, but soldiers’ ovarioles were small and lacking gastrulating embryos. We also found that soldier reproduction is constrained by apoptosis in maternal nutritive cells and necrosis in oocytes and embryos.
C. japonica also contains endosymbionts. The eusocial aphid
C. japonica provides us with a unique opportunity to study how symbiosis operates in a social setting. We are currently investigating the interface of symbiosis and sociality.







