The medaka is a small egg-laying fresh water fish found in brooks and rice paddies in Eastern Asia. This species has a long history as an experimental animal, especially in Japan. Our laboratory has conducted studies on the dynamics of transcriptional and chromatin accsssibility landscape during medaka development, the understanding of pigment cell differentiation using body color mutants, genome sequence of the Javanese medaka, Oryzias javanicus and the molecular genetic basis of diversified sexually dimorphic traits in Oryzias species, In addition to these activities, our laboratory was charged with the responsibility of leading the National BioResource Project Medaka (NBRP Medaka) from 2007.
I. Daynamic transcriptional and chromatin accesibikity landscape during medaka development.
A high-quality genome sequence and a variety of genetic tools are available for medaka. However, existing genome annotation is still rudimentary, as it was mainly based on computational prediction and short-read RNA-seq data. To overcome this situations, we conducted long-read RNA-seq, short-read RNA-seq, and ATAC-seq coraboration with Dr. Tu at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences. This work constructed a much-improved gene model set including about 17,000 novel isoforms and identified 1600 transcription factors, 1100 long non-coding RNAs, and 150,000 potential cis-regulatory elements as well. The work provides the first comprehensive omics datasets of medaka embryogenesis. The data portal (
http://tulab.genetics.ac.cn/medaka_omics) will serve as a daily reference tool for the entire medaka community.
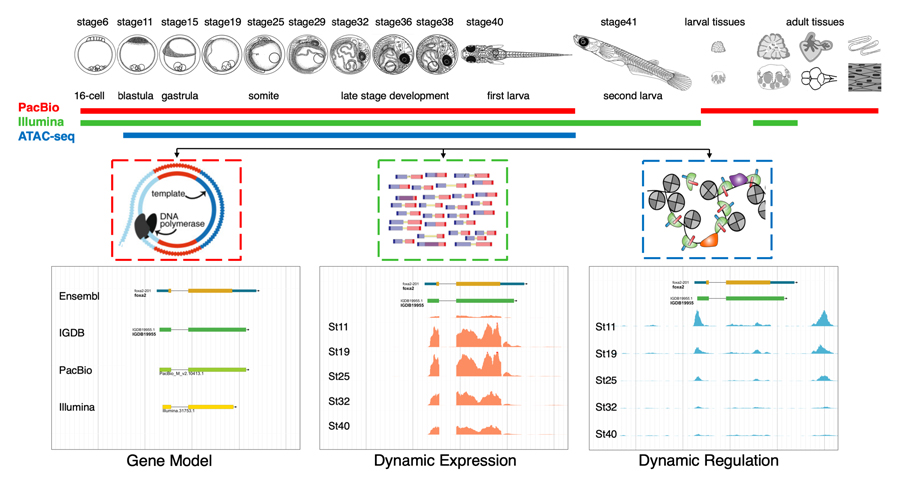
Figure 1. Overview of medaka multi-omics anslysis during development
II. The Medaka Inbred Kiyosu-Karlsruhe (MIKK) panel and their genomic variation and epigenomic landscape
The Medaka Inbred Kiyosu-Karlsruhe (MIKK) panel is the first near-isogenic panel of 80 inbred lines derived from a wild founder population collected from Kiyosu, Toyohashi. Inbred lines provide fixed genomes that make it possible to repeat studies, to vary both genetic information and environmental conditions in controlled studies, as well as to conduct functional studies. MIKK will thus make it possible to investigate phenotype-to-genotype association studies of complex genetic traits while carefully controlling interacting factors. Its applications are numerous, and include genetic research, human health, drug development, and fundamental biology. Using short reads from Illumina, we further characterise each of the 80 MIKK lines according to their genetic composition, using the Southern Japanese medaka inbred strain HdrR as a reference genome. We used Oxford Nanopore long read sequencing technology to analyze 12 representative lines from MIKK to analyze the larger and more complex structural variations. We also investigate line-specific CpG methylation and performed differential DNA methylation analysis across these 12 lines.
III. Genome sequence of the Javanese medaka, Oryzias javanicus, as a model for studying seawater adaptation
Medaka fish in the genus
Oryzias are an emerging model system for studying the molecular basis of vertebrate evolution. This genus contains approximately 35 species and exhibits great morphological, ecological and physiological differences among it’s species. Among these species, the Java medaka,
Oryzias javanicus, is the species that has most typically adapted to seawater. We sequenced and assembled the whole genome of
O. javanicus, as a model fish species for studying molecular mechanisms of seawater adaptation. In teleost fish, the major osmoregulatory organs are the gills, intestine and kidney, and these play different roles to maintain body fluid homeostasis. Many genes encoding hormones, receptors, osmolytes, transporters, channels and cellular junction proteins are potentially involved in this osmotic regulation. In addition to the osmoregulation, hatching enzyme activity dramatically changes in different salt conditions. At the hatching stage, fish embryos secrete a specific cocktail of enzymes in order to dissolve the envelope. In the medaka
O. latipes, digestion of the envelope occurs after the cooperative action of two kinds of hatching enzymes: (i) the high choriolytic enzyme (HCE) and (ii) the low choriolytic enzyme (LCE) (Yasumasu
et al., 2010).The HCE shows higher activity in freshwater than in brackish water (Kawaguchi
et al., 2013). Thus, availability of the high-quality reference genome in
O. javanicus would facilitate further research for investigating the molecular basis of physiological differences including the osmotic regulation and the hatching enzyme activity among
Oryzias species.
IV. National BioResource Project Medaka
Our laboratory has been acting as main facility of the Medaka BioResource Project (NBRP Medaka) in the fourth phase of NBRP, which started in 2017. We are providing wild strains, and related species, genomic resources (approximately 400,000 cDNA clones from 30 different cDNA libraries (containing approximately 23,000 different sequences) and BAC/Fosmid clones covering the entire medaka genome), and hatchery enzymes essential for embryo manipulation and live imaging. These bioresources are available on our website (Figure 3. https://shigen.nig.ac.jp/medaka/). These bioresources can be searched by various methods such as keywords, sequence homology, and expression profiles using the database on the website. In addition, genome browsers for the three inbred strains, phylogenetic relationships between wild strains of medaka and related species, and experiment manuals are also available. With the approval of the second supplementary budget for FY2020, we were able to install a cabinet-type fish tank washing machine. This has freed up our technical support staff from the need to wash the tanks by hand, and has allowed us to focus more on breeding and management, which requires more human work. In addition, a system to remotely monitor the temperature, humidity, and illumination in the medaka breeding rooms and the water temperature in the breeding tanks was installed. The air conditioner in the breeding room was also upgraded. We have continuously monitored the medaka breeding conditions using these systems.

Figure 2. NBRP Medaka website