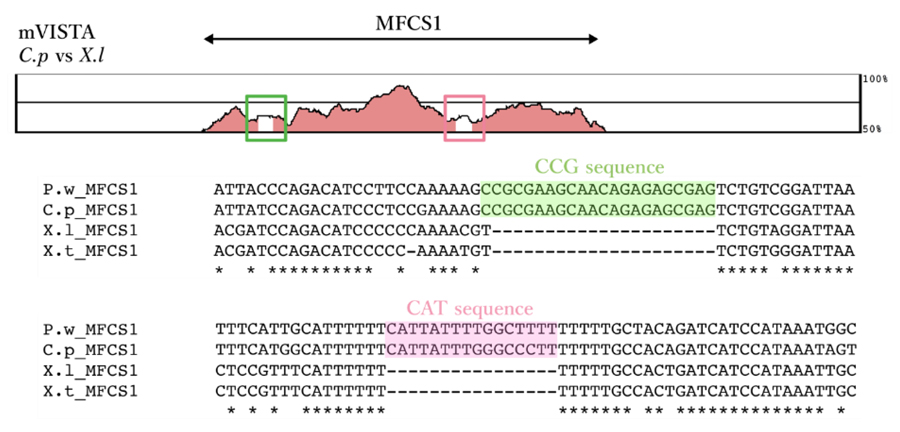
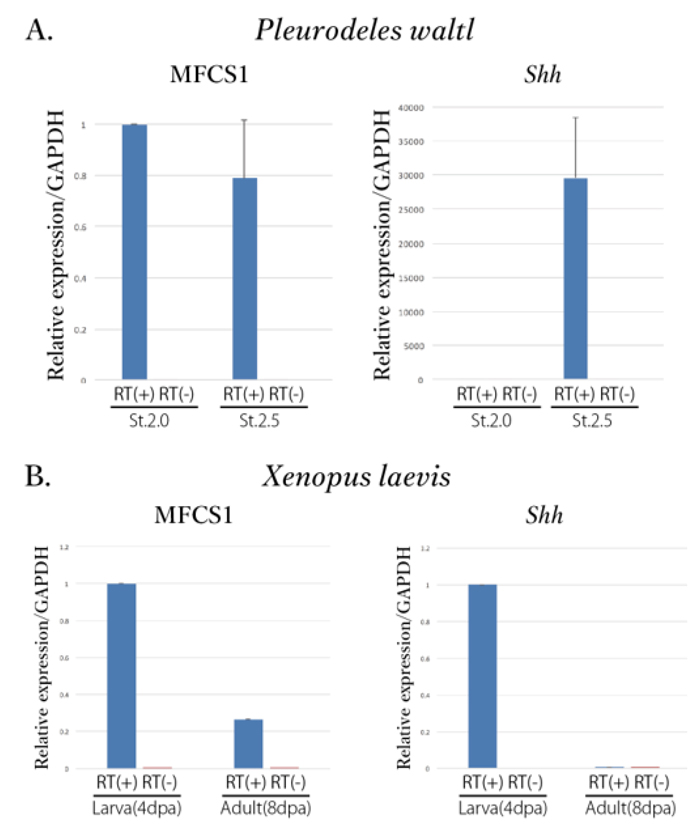
Satoh, A., Kashimoto, R., Ohashi, A., Furukawa, S., Yamamoto, S., Inoue, T., Hayashi, T., Agata, K. (2022). An approach for elucidating dermal fibroblast dedifferentiation in amphibian limb regeneration. Zoological Lett.
8, 6. doi:
10.1186/s40851-022-00190-6.
Sato, Y., Shibata, N., Hashimoto, C., Agata, K. (2022). Migratory regulation by
MTA homologous genes is essential for the uniform distribution of planarian adult pluripotent stem cells. Dev. Growth Differ.
64, 150-162. doi:
10.1111/dgd.12773. Epub 2022 Feb 28.
Takeuchi, T., Matsubara, H., Minamitani, F., Satoh, Y., Tozawa, S., Moriyama, T., Maruyama, K., Suzuki, K.T., Shigenobu, S., Inoue, T., Tamura, K., Agata, K., Hayashi, T. (2022). Newt
Hoxa13 has an essential and predominant role in digit formation during development and regeneration. Development
149, dev200282. doi:
10.1242/dev.200282. Epub 2022 Mar 11.
Lee, H., Hikasa, K., Umesono, Y., Hayashi, T., Agata, K., Shibata, N. (2022). Loss of
plac8 expression rapidly leads pluripotent stem cells to enter active state during planarian regeneration. Development
149, dev199449. doi:
10.1242/dev.199449. Epub 2022 Feb 3.