
National Institute for Basic Biology





Although transposons occupying a large portion of the genome in various plants were once thought to be junk DNA, they play an important role in genome reorganization and evolution. Active DNA transposons are important tools for gene functional analysis. The endogenous non-autonomous transposon, nDart1, in rice (Oryza sativa L.) is said to generate various transposon-insertion mutants because nDart1 elements tend to insert into genic regions under natural growth conditions. The transpositions of nDart1 were promoted by an active autonomous element, aDart1-27, on chromosome 6. By using the endogenous nDart1/aDart1-27 system in rice, a large-scale nDart-inserted mutant population was easily generated under normal field conditions, and the resulting tagged lines were free of somaclonal variation. The nDart1/aDart1-27 system was introduced into a rice variety, Koshihikari, named MK-1. 3000 MK-1 plants were grown in field conditions (IPSR, Okayama Univ.). All plants’ genomes were isolated for identifying the insertion sites of nDart1.
Seed size and number were controlled by various genes in the plants. It was reported that expression changes in high contribution genes for seed size, number and panicle shape resulted in a decrease of the total yield. A strategy for boosting rice yield based on molecular biology is to stack the finely tuned gene expressions. The Lgg mutant which was isolated from MK-1 plants bore slightly larger grains (Figure 1) as a dominant inheritance. Transposon-display identified the insertion site of nDart1 in the Lgg mutant.

Figure 1. Phenotype of Large grain (Lgg). Harvested panicle and seeds.
The identified LGG gene shows similarity to RNA binding proteins. In the Lgg mutants, insertion of nDart1 and genomic deletion was confirmed in a 5’ untransrated region of the LGG gene. It was estimated that genomic deletion in Lgg mutants derived from microhomology in both the nDart1 and LGG-genomic region. The expression of the LGG gene in Lgg mutants was lower than WT. Lgg mutation was essentially caused by the insertion of nDart1, knock-out mutant using genome editing (GE) and over-expressing (OE) lines were generated (Figure 2). GE lines showed large grain phenotypes that were the same as Lgg mutants. On the contrary, harvested seeds from the OE line were smaller than the WT and GE lines. To confirm that Large grain in Lgg mutants was caused by increasing the cell size or cell number or both, we developed a cross section method of rice seeds by using cryomicrotome (Chiou et al. 2018).
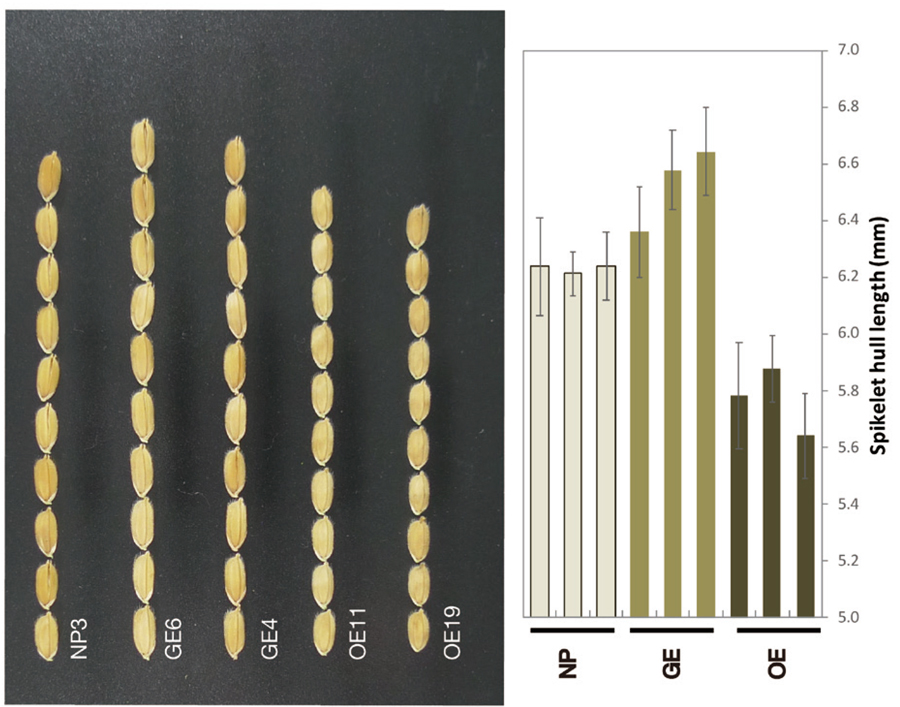
Figure 2. Phenotypes of wild type (NP), the genome-editing line (GE), and the overexpression line (OE). (Left)The spikelets. Bar = 5 mm. (Right) Spikelet hull length of NP, GE) and OE plants. Different letters indicate P < 0.05 by Fisher’s Least Significant Difference test. n = 10. mean ± SD.