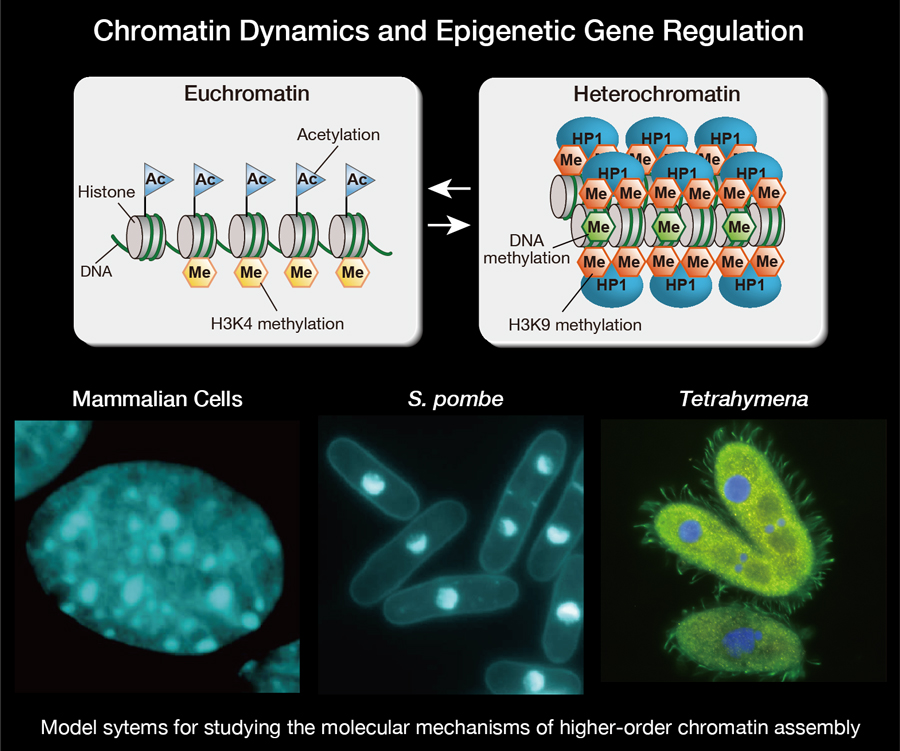
Research Summary
The stable inheritance of gene expression or repression states is essential to cellular differentiation and development, and an epigenetic cellular-memory system derived from higher-order chromatin structure plays a fundamental role in this process. The assembly of higher-order chromatin structures has been linked to the covalent modifications of histone tails, chromatin-associated proteins, and non-coding RNAs. However, the exact means by which this chromatin-based epigenetic information is established and faithfully maintained across cell divisions and throughout development remains unclear. To try to gain a better understanding of the molecular mechanisms underlying chromatin-based epigenetic phenomena, our lab uses model systems including mammalian culture cells, fission yeast, Schizosaccharomyces pombe (S. pombe), and ciliate Tetrahymena for studying the assembly of higher-order chromatin structures, known as heterochromatin. We are also attempting to determine the cellular functions of chromatin modifying factors so that we can develop a better understanding of complicated epigenetic phenomena in higher eukaryotic cells.
Research Projects
-
• Revealing the regulation mechanisms of histone modifying enzymes
-
• Characterizing the roles of HP1 family proteins in assembling higher-order chromatin structures
-
• Analysis of chromatin dynamics in dormant cells
I. Establishment and maintenance of higher-order chromatin structures
1-1 Mechanisms regulating Clr4 histone methyltransferase activity
In eukaryotic cells, the assembly of heterochromatin plays an important role in diverse chromosomal processes and epigenetic gene regulation. Heterochromatin is characterized by the methylation of histone H3 at lysine 9 (H3K9me). H3K9me is catalyzed by SUV39H-family histone methyltransferases, and functions as a binding site for recruiting heterochromatin protein 1 (HP1) family proteins. In fission yeast, H3K9me is catalyzed by histone methyltransferase Clr4. Clr4 has two functional domains, the N-terminal chromodomain (CD), which recognizes H3K9me, and the C-terminal SET domain responsible for Clr4’s enzymatic activity (Figure 1A). Since Clr4’s uncontrolled activity leads to inappropriate H3K9me and aberrant gene silencing, its enzymatic activity needs to be strictly controlled. However, the detailed mechanisms regulating Clr4’s activity are poorly understood.
A previous study showed that N-terminally deleted Clr4 mutant exhibits a stronger activity than full-length Clr4, suggesting that the Clr4 N-terminal region negatively regulates the activity of C-terminal SET domain. To examine the regulation mechanisms of Clr4’s activity, we tested whether Clr4 N-terminal region interacts with its C- terminal SET domain. Pull-down assays using recombinant proteins reveal that the Clr4 N-terminal region physically interacts with the SET domain and that this interaction requires both CD and its adjacent region. To determine the regions/residues responsible for this interaction, we performed crosslinking mass spectrometry (Figure 1B) and found that crosslinked residues were concentrated in a region near the C-terminal end (Figure 1C). Interestingly, mutant Clr4 proteins containing amino-acid substitutions at this region exhibit higher methyltransferase activity. These results suggest that Clr4’s enzymatic activity is negatively regulated by intramolecular interactions.
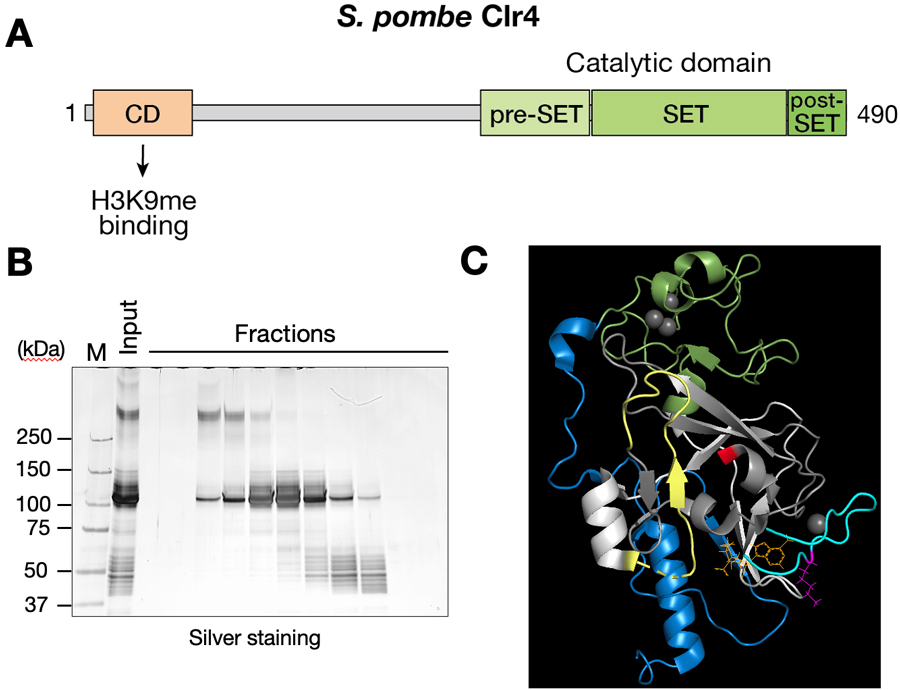
Figure 1. Characterization of intramolecular interactions of Clr4. (A) Domain organization of Clr4. The N-terminal chromodomain (CD) recognizes H3K9me, and the C-terminal SET domain is responsible for Clr4’s enzymatic activity. (B) Crosslinking mass spectrometry. After in vitro crosslinking reaction, recombinant Clr4 proteins were separated by gel-filtration chromatography and analyzed by SDS-PAGE and silver staining. (C) Residues identified in the cross-linking mass spectrometry to be involved in the interaction are mapped on 3D structure of Clr4 SET domain.
II. Roles of HP1 family proteins in assembling higher-order chromatin structure
2-1 Characterizing the role Chp2 in heterochromatin assembly in fission yeast
Heterochromatin protein 1 (HP1) is an evolutionarily conserved chromosomal protein that binds H3K9me, and plays a crucial role in forming higher-order chromatin structure. HP1 family proteins share a basic structure consisting of an N-terminal CD and a C-terminal chromoshadow domain (CSD) linked by an unstructured henge region (Figure 2A). The CD functions as a binding module that target H3K9me, whereas CSD functions as a dimerization module that provides a recognition surface to bind and recruit other proteins. In fission yeast, two HP1 family proteins, Swi6 and Chp2, are involved in heterochromatic silencing. While Swi6 is expressed abundantly and plays a dose-dependent role, Chp2 is expressed at a lower level and displays a novel ability to tightly bind chromatin-enriched fraction independently of H3K9me. To dissect how this Chp2-specific ability contributes to its function, we performed Tandem Affinity Purification (TAP) to identify Chp2-specific binding partners and successfully identified novel candidates for Chp2 binding partners. Furthermore, we examined Chp2-specific DNA-binding activity by Electrophoretic Mobility Shift Assay (EMSA) and found that both the hinge and CSD neighboring regions make distinct contribution to its DNA binding. These results suggest that both the Chp2’s property to bind specific binding partners and its intrinsic DNA binding ability contribute to its tight chromatin association and also to division of labor between Swi6 and Chp2.
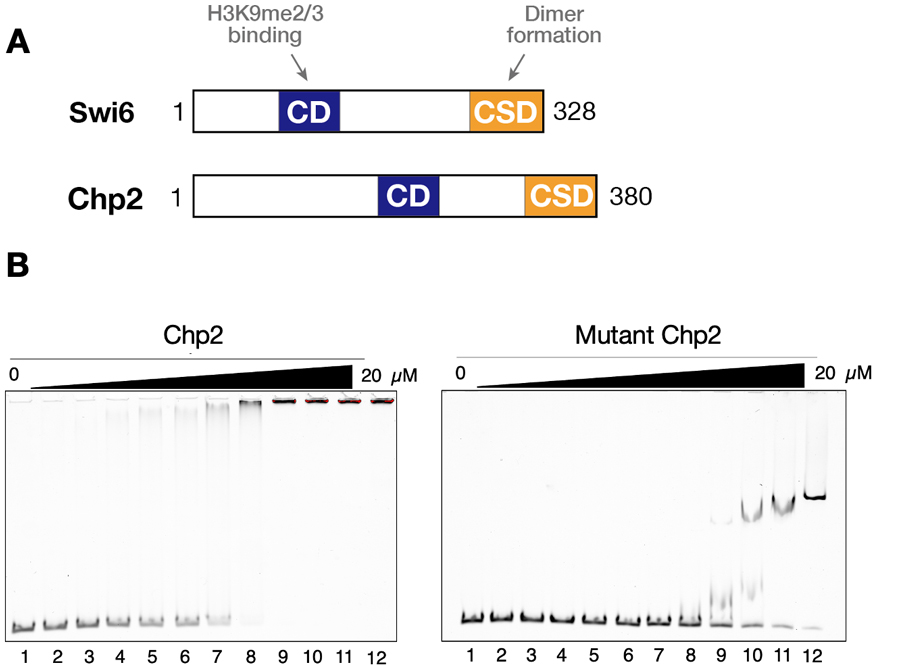
Figure 2. Chp2 possesses a strong DNA/RNA-binding activity. (A) Schematic diagram of Swi6 and Chp2. The chromodomain (CD) and the chromoshadow domains (CSD) are shown. (B) Representative results of EMSAs performed with control and mutant Chp2.
2-2 Multiple HP1-like protein-containing complex regulates DNA elimination in Tetrahymena
Heterochromatin plays important roles in transposon (TE) silencing. A major type of heterochromatin contains chromatin, that is histone H3 methylated at lysine 9/27 (H3K9/27me), and its reader HP1 proteins, that recruit diverse proteins onto the chromatin to silence the TEs. Although multiple HP1 proteins are co-expressed in many eukaryotic cells, the interplay between these HP1 proteins has been elusive. Here, we show that subset of the HP1 proteins form a complex and play important roles to eliminate TE-related Internal Eliminated Sequences (IESs) from the somatic genome during macronuclear development in the ciliated protozoan Tetrahymena. We tethered 7 HP1-like proteins individually to the artificially created locus by the LexA-LexO system and found that only 4 of them including Pdd1 induced the elimination of the tethered site. This ectopic DNA elimination was exclusively achieved by their chromoshadow domains (CSDs), indicating that the CSDs of the distinct type of HP1-like proteins recruits all the proteins that are required for DNA elimination. Immunoprecipitation of Pdd1 specifically enriched the other HP1-like proteins that were sufficient for the ectopic DNA elimination. The chromodomains of a subset of HP1-like proteins showed strong affinity to both H3K9me3 and H3K27me3. Overall, these results suggest that multiple HP1-like proteins cooperatively recognize the methylated histones and form core complex to recruit other effector proteins for DNA elimination (Figure 3).
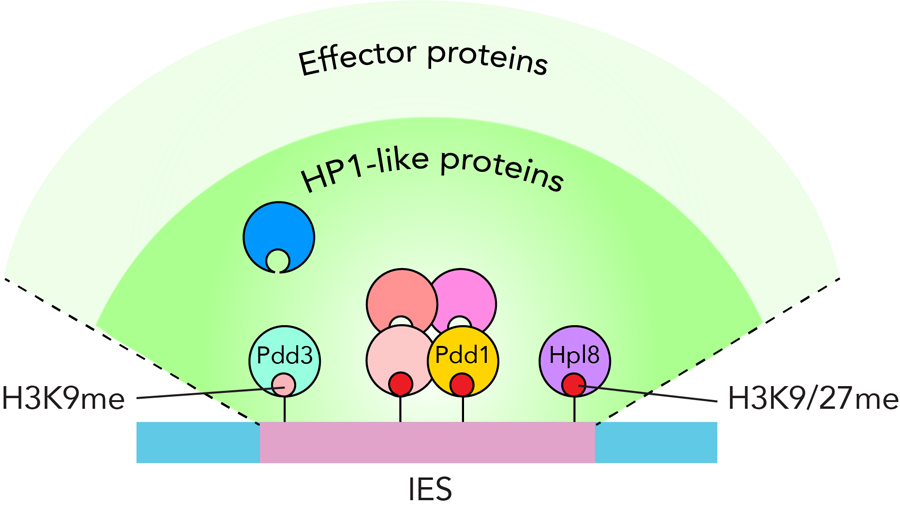
Figure 3. Model for how multiple HP1-like protein-containing complex regulates DNA elimination in Tetrahymena.
2-3 The HP1-like protein Hpl8p is important for programmed DNA elimination in Tetrahymena
Transposon mobility represents a threat for the genome integrity in all living things. To counter this, the ciliated protozoan Tetrahymena uses heterochromatin to selectively eliminate over 10,000 transposon loci from its somatic nucleus in the developmentally programmed process of DNA elimination. To address how this process is coordinated by heterochromatin components, we focus on an HP1-like protein named Hpl8p that localizes to the developing macronucleus during heterochromatin formation and DNA elimination (Figure 4). The chromodomain of Hpl8p recognized both H3K9me3 and H3K27me3 in vitro. HPL8 disrupted mutants showed defect in vegetative growth and in the progression of sexual reproduction. FISH analyses using a probe against one of the transposons showed that more than 90% of HPL8 disrupted cells did not complete its elimination, suggesting that Hpl8p plays important roles in DNA elimination. Altogether, these results suggest that Hpl8p binds the H3K9/27 methylation marks over transposon loci and facilitates the heterochromatin formation by interacting with heterochromatin components required for DNA elimination. Furthermore, we propose that Hpl8p also plays important roles in suppression of unwanted genes during the vegetative growth and sexual reproduction process possibly by recognizing H3K27me3 in the transcriptionally active macronucleus.
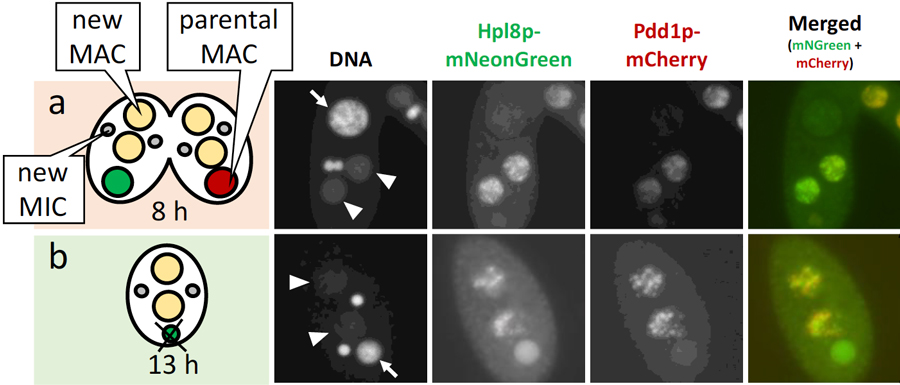
Figure 4. Cells expressing the Hpl8-mNeonGreen (green) and Pdd1p-mCherry (red) at 8 and 13 hours post induction of mating are shown in (a) and (b), respectively. DNA was counterstained with DAPI. Arrows and arrowheads indicate the parental and new MACs, respectively.
III. Analysis of chromatin dynamics in spore nuclei of fission yeast
Gametogenesis is a crucial process for sexually reproducing organisms to produce haploid gametes from diploid cells. Generally, epigenetic memory is erased from zygote nucleus, a phenomenon known as reprogramming, resulting in increased totipotency. On the other hand, it has been demonstrated in some organisms that a part of epigenetic memory can be inherited transgenerationally. The balance between reprogramming and epigenetic inheritance plays a key role in the developmental processes of offspring. It has been also reported that some gametes, like plant sperm and mammalian spermatozoa, have highly compacted nuclei and have altered histone modifications. However, the mechanisms underlying how epigenetic marks are regulated during gametogenesis remain to be fully elucidated. To gain an insight into epigenetic inheritance and chromosome structural change during gamete formation, we analyzed chromatin dynamics in spore nucleus of fission yeast (Figure 5A). We isolated fission yeast spores by density-gradient centrifugation and performed mass spectrometry analysis to determine spore-specific nuclear proteins (Figure 6B). We further examined the change in histone modifications of spore nuclei. Chromatin immunoprecipitation analysis revealed that the levels of H3K9me at heterochromatic regions had not been noticeably changed, but the levels of H3K4me associated transcriptionally active regions had clearly increased in spore nuclei. We also found that H3K4me distribution at coding regions in spore genome clearly different from that of control vegetative cells. These results imply that dynamic changes in active histone marks contribute to spore formation and epigenetic inheritance.
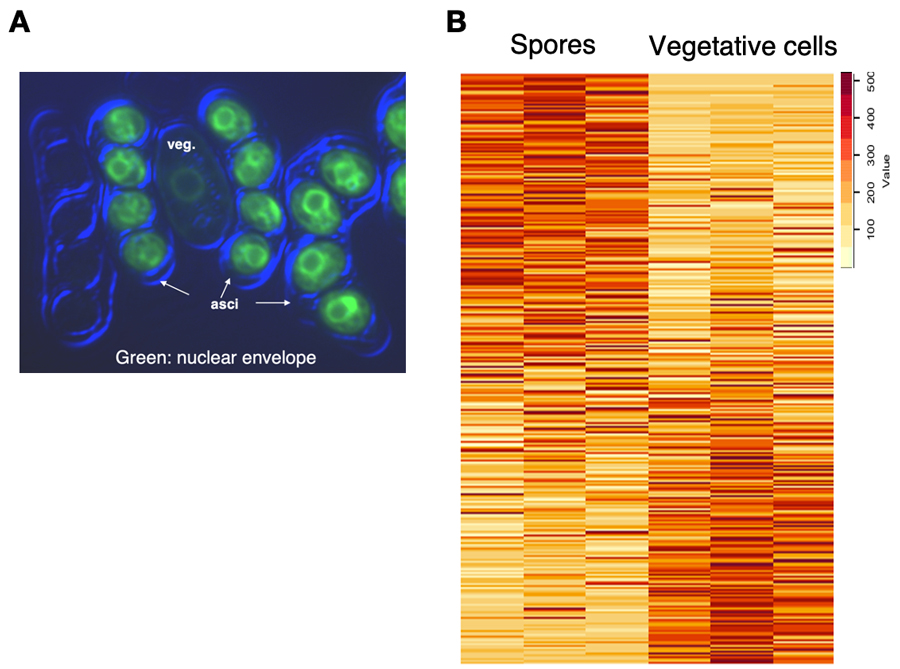
Figure 5. Analysis of chromatin dynamics in spore nuclei of fission yeast. (A) A representative image of vegetative cells (veg.) and spores in asci. (B) Heatmap representation of proteins detected in spores and vegetative cells.
Selected Publications
Ding, D.Q , Okamasa, K., Yoshimura, Y., Matsuda, A., Yamamoto, T.G., Hiraoka, Y., and Nakayama, J. (2024). Proteins and noncoding RNAs that promote homologous chromosome recognition and pairing in fission yeast meiosis undergo condensate formation in vitro. FASEB J. 38, e70163.
Kawaguchi, T., Hashimoto, M., Nakagawa, R., Minami, R., Ikawa, M., Nakayama, J., and Ueda, J. (2024). Comprehensive posttranslational modifications in the testis-specific histone variant H3t protein validated in tagged knock-in mice. Sci Rep. 14, 21305.
Rahayu, A.F., Hayashi, A., Yoshimura, Y., Nakagawa, R., Arita, K., and Nakayama, J. (2023). Cooperative DNA-binding activities of Chp2 are critical for its function in heterochromatin assembly. J. Biochem. 174, 371-382.
Dong, C., Nakagawa, R., Oyama, K., Yamamoto, Y., Zhang, W., Dong, A., Li, Y., Yoshimura, Y., Kamiya, H., Nakayama, J., Ueda, J., and Min, J. (2020). Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19. eLife 9, e58675.
Ding, D.Q., Okamasa, K., Katou, Y., Oya, E., Nakayama, J., Chikashige, Y., Shirahige, K., Haraguchi, T., and Hiraoka, Y. (2019). Chromosome-associated RNA-protein complexes promote pairing of homologous chromosomes during meiosis in Schizosaccharomyces pombe. Nat. Commun. 10, 5598.
Oya, E., Nakagawa, R., Yoshimura, Y., Tanaka, M., Nishibuchi, G., Machida, S., Shirai, A., Ekwall, K., Kurumizaka, H., Tagami, H., and Nakayama, J. (2019). H3K14 ubiquitylation promotes H3K9 methylation for heterochromatin assembly. EMBO Rep. 20, e48111.











