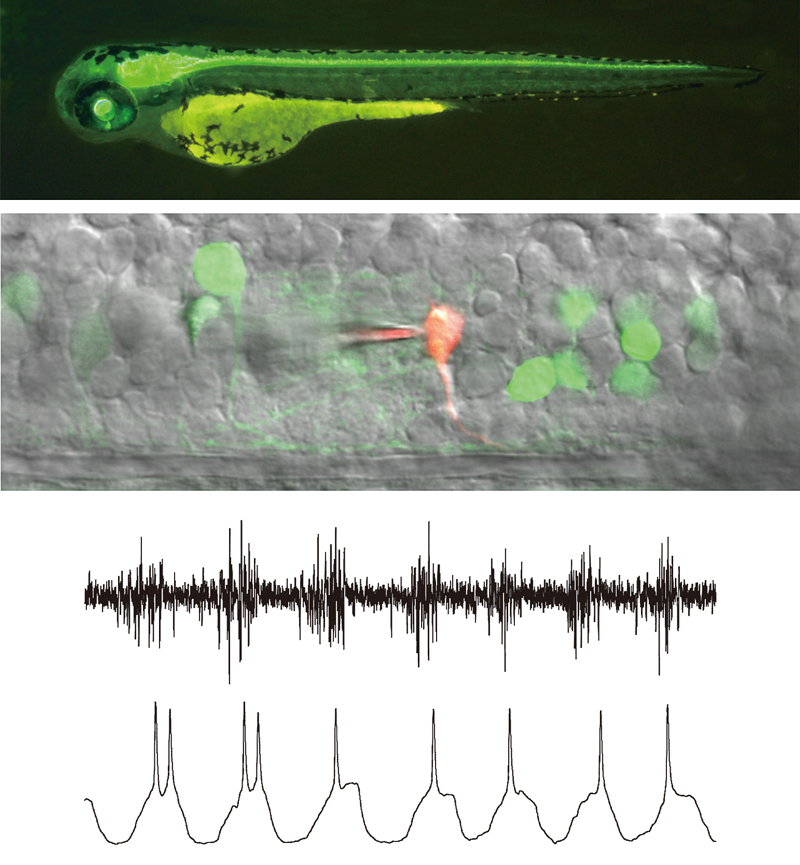
The elucidation of how neural circuits in the central nervous system operate at the resolution of single neurons during behavior is one of the major goals of neuroscience. Our research division is working to uncover the operational mechanisms of the motor systems in the spinal cord and brainstem during the generation of behavior, using larval zebrafish, a small fish species with a simple central nervous system. A key technique in our approach is the creation of transgenic fish, which allows us to visualize specific classes of neurons in living organisms. This enables us to perform calcium imaging and electrophysiological analyses on the neurons in question. Recently, we have been actively advancing research on the structure and operational mechanisms of neural circuits involved in posture control, using a custom microscope equipped with an electric rotating stage that we developed in-house.
Research Projects
-
• Neuronal circuits that control rhythmic pectoral fin movements
-
• Mechanism for detecting vestibular information in the otolith organ
-
• Neuronal circuits for postural control
The vertebrate central nervous system (CNS) contains many different types of neurons that form at distinct characteristic positions, and develop specific axonal connections and functions. This complexity has made it difficult to perform detailed functional analysis of neuronal circuits. In particular, it has been very difficult to reproducibly identify cell types. However, molecular genetic studies conducted over the past 15 years have strongly suggested that the expression of transcription factors in the developing CNS helps determine the morphological and functional properties of neurons. This has opened up the possibility that researchers can use these transcription factors as markers to identify cell types in the CNS. Transgenic animals that express fluorescent protein in specific subsets of neurons are particularly powerful tools in studying the functions of the corresponding neurons in the neuronal circuits.
To fully exploit the methodology described above, we use larval zebrafish as experimental animals. The biggest advantage of doing so is that larval zebrafish are almost completely transparent. This allows us to utilize many optical techniques, including morphological/functional imaging and optogenetics. We can also perform targeted in vivo electrophysiological recording with relative ease using this transparent model. An additional advantage of zebrafish is that their CNS is much simpler than that of mammals. This enables us to perform detailed functional analysis of neuronal circuits at a single cell resolution. Our hope is to reveal the operational principles of vertebrate CNS by using this simple system.
We have been focusing on studying neuronal circuits that control locomotion. Much of the control of locomotor movements is accomplished by neuronal circuitry located in the spinal cord. Therefore, the focus of our studies has been spinal neuronal circuits in larval zebrafish.
In addition to zebrafish, we have also started to use medaka as experimental animals. Medaka have many advantages that are similar to those of zebrafish Because NIBB is the main hub of the Medaka National Bioresource Project, we are ideally located in regards to experiments using medaka. To begin with, we explored whether knock-in fish could be efficiently generated using the CRISPR/Cas9 technique.
I. Generation of Transgenic zebrafish
We have been generating transgenic zebrafish that express fluorescent proteins (GFP or RFP), Gal4, or Cre in specific classes of neurons in the CNS by using gene promoters/enhancers of genes and are known to be expressed in subsets of neurons. Most of the genes we used are transcription factors expressed in subsets of neurons in the developing CNS. We also used genes whose expressions are tightly related to neurotransmitter properties of neurons (i.e., vesicular glutamate transporter).
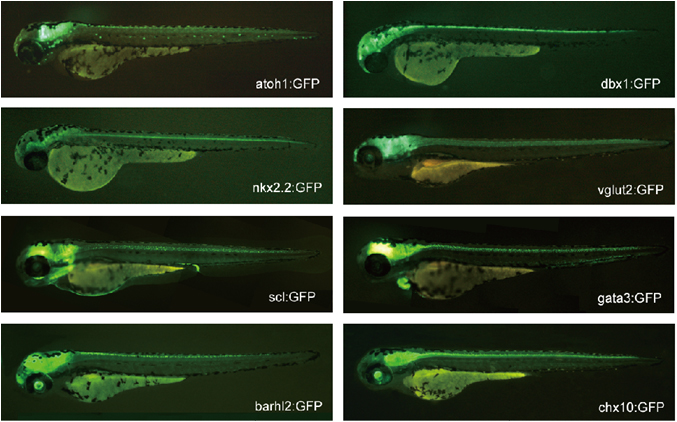
Figure 1. Examples of transgenic fish expressing GFP in specific classes of neurons.
In our early studies, we used a BAC-based transgenic technique for the generation of transgenic fish. By 2014, we succeeded in establishing a reliable knock-in method by utilizing the CRISPR-Cas9 system. The method we have developed is highly efficient, so much so that nearly one-third of the animals we raise become transgenic founders. Thus far, we have established more than 20 knock-in transgenic fish. Thus, this method greatly facilitates our functional analysis on neuronal circuits.
II. Long descending commissural V0v neurons ensure coordinated swimming movements along the body axis in larval zebrafish
In the early developmental stage, most animals can only exhibit immature forms of behaviors. As development progresses, they acquire the ability to produce more mature or refined forms of behaviors. Concurrent with such changes, many new connections are formed in the nervous system, which suggests that the formation of new connections is linked to the developmental maturation of behaviors. In animals in which new neurons are generated during development (e.g., fish and amphibians), the incorporation of new neurons into the pre-existing circuits together with the forming of new connections likely contributes to the maturation of movements.
One example of this developmental maturation of behaviors is seen in the swimming behavior of larval zebrafish. Older larvae (i.e., 4–5 dpf) exhibit more refined forms of swimming than younger larvae (i.e., 2 dpf), During this period (from 2 dpf to 4–5 dpf), new neurons are generated in both the brain and spinal cord, with the latter being mainly responsible for generating swimming outputs. One class of premotor spinal neurons that are added later to the early spinal neuronal circuits are MCoD neurons, a subclass of V0v neurons (V0v neurons represent an excitatory class of neurons derived from the p0 developmental domain of the spinal cord). MCoD neurons are absent in the early stages and develop in a later phase of neurogenesis in the spinal cord. MCoD neurons are active during slow swimming. As for their function, one study showed that they contribute to the general excitability of spinal swimming circuits, and their ablation decreased the occurrence frequency of spontaneous swimming. Their function in the more specific aspect of slow swimming remains elusive, however.
One of the most characteristic features of slow swimming in older larvae is the stability of the head in the yaw dimension. In slow swimming, the muscle contractions are mostly confined to the trunk that is caudal to the swim bladder. Given that the center of the mass is located near the swim bladder in larval zebrafish (Figure 2A), it is thought that the head yaw displacement is produced by the recoil of the yawing moment force generated in the trunk. Considering this, the stability of the head yaw indicates that the net yawing moment force in the trunk that acts to the center of the mass is very small during slow swimming. Swimming consists of a descending wave of muscle contraction along the trunk. With this movement, the bending of the body transmits force to the surrounding water, and this region of the body, in turn, receives reaction force. To make the net yawing moment force minimal, the movements of the rostral and caudal parts of the trunk need to be highly coordinated with the diagonal dimension; when the rostral part receives leftward force, for example, the caudal part needs to receive rightward force. For this to occur, the swimming body form cannot be C-shaped (unilateral body bend); rather, the shape needs to be sinusoidal (S-shaped). MCoD neurons are a good candidate for implementing this coordinated movement of the trunk in the diagonal dimension, because they are active during slow swimming, and because they are long-distance descending commissural excitatory neurons that make direct connections onto MNs in the caudal region of the contralateral spinal cord (Figure 2B).
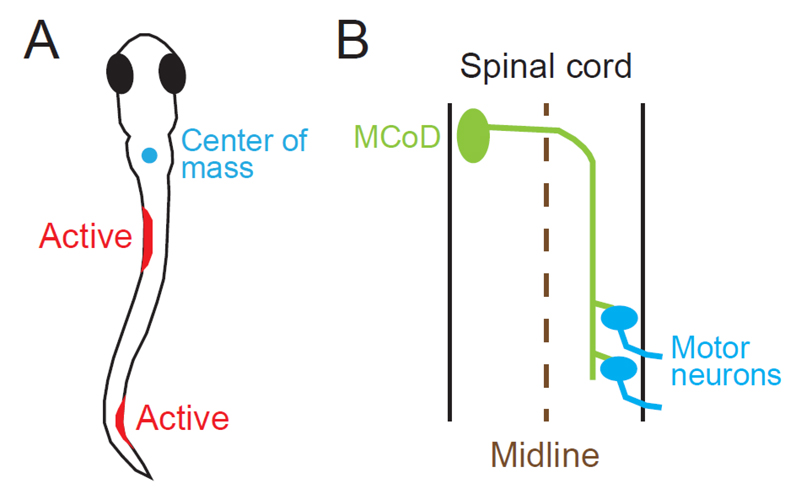
Figure 2. (A) One of the swim forms of a zebrafish larva at 4 to 5 dpf. The cyan circle shows the center of mass, which is located near the swim bladder. Muscle contractions are presumed to occur in the two locations marked in red. (B) Projection pattern of an MCoD neuron in the spinal cord. The axon of the MCoD neuron crosses the midline (broken line), descends on the contralateral spinal cord, and makes mono-synaptic excitatory connections onto caudally located MNs.
In this study, we tested whether MCoD neurons play a role in the coordinated movements of the trunk in the diagonal dimension, thereby ensuring minimal head yaw displacement during slow swimming. Our laser ablation experiments revealed that MCoD neurons do indeed play the expected role. In the MCoD-ablated larvae, the normal S-shaped body form during swimming was often lost with increased appearance of unilateral C-shaped bends. Concurrently, the head yaw stability was greatly impaired (Figure 3). In addition to swimming, the present study also sheds light on the evolutionarily conserved role of V0v neurons. In mice, long-distance descending commissural V0v neurons have been implicated in interlimb coordination during walking in the diagonal dimension18. We suggest that the long-distance descending commissural V0v neurons for the coordinated movements of the body/limbs in the diagonal dimension are the evolutionarily conserved pathway in spinal locomotor circuits.
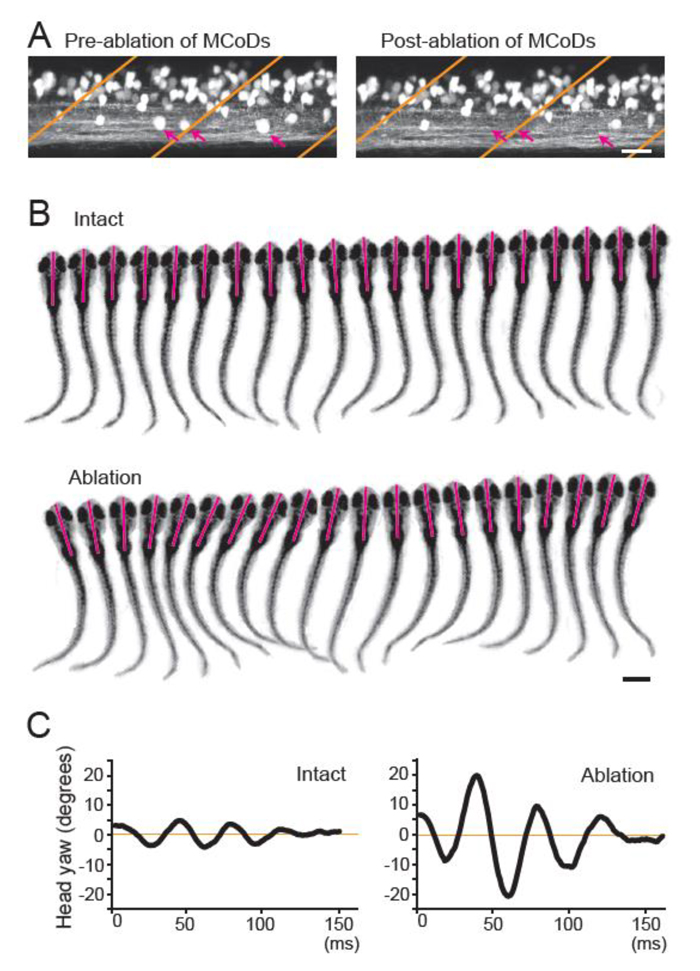
Figure 3. (A) Confocal stacked images of Tg[evx2-hs:GFP] fish before (left) and after (right) laser ablation. Images of two hemi-segments are shown. Magenta arrows show MCoD neurons that were chosen for laser ablation. MCoD neurons can be identified by their very ventral location in the spinal cord. Brown lines show boundaries of muscle segments. Scale bar, 20 μm. (B) Successive images captured at 1000 frames per second of larval zebrafish swimming. Images of every three frames (3 ms interval) are shown. Magenta bars depict the head directions in each frame. Top, images of an intact fish. Bottom, images of an MCoD-ablated fish. Scale bar, 500 μm. (C) Graphs of head yaw angle (y axis) versus time (x-axis) during swimming. Left, intact fish. Right, MCoD-ablated fish.
III. In vivo functional imaging analysis of the vestibular sensory organ
Maintaining head and body orientation relative to the Earth’s vertical gravity axis is vital for survival. Vestibular organs in the inner ear play a crucial role for this task. Sensory hair cells in the otolith organs receive linear acceleration, e.g., head tilt, translation and vibration. Direction of the acceleration is detected by the polarized arrangement of hair bundles in the hair cells (Figure 4). Each otolith organ contains hair cells with different but topographically organized hair-bundle polarity that reverses at a line of polarity reversal (LPR). Although morphofunctional specialization of the vestibular hair cells has been widely studied, the direction- and modality-selective responses to the head motion have not been systematically studied in vivo, therefore how the head motion signals are processed in the vestibular system remains unclear.
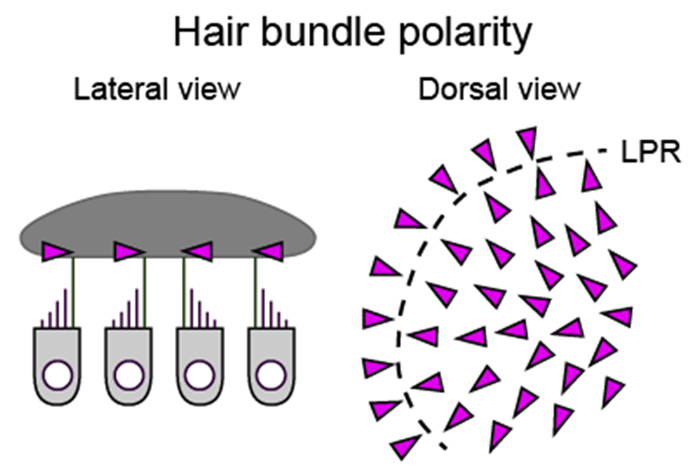
Figure 4. Hair bundle polarity (arrowheads) in the utricle.
To visualize hair cell responses to the head motion, we built a microscope in which an objective lens can tilt with a small sample 360 degree by a motorized stage during Ca2+ imaging (Figure 5). A spinning-disk confocal scanner and an image splitting optics formed green and red fluorescent images on a digital camera. This ratiometric imaging setup reduced artifacts derived from non-uniformity of the excitation light and optical distortion during the optics motion. With this tiltable objective microscope, we imaged neural activity in all the utricular hair cell at the single-cell level during pitch or roll tilt/vibration in 5-day-old transgenic zebrafish larvae expressing Ca2+ indicator, jGCaMP7f, and red fluorescent protein, tdTomato, in the hair cells.
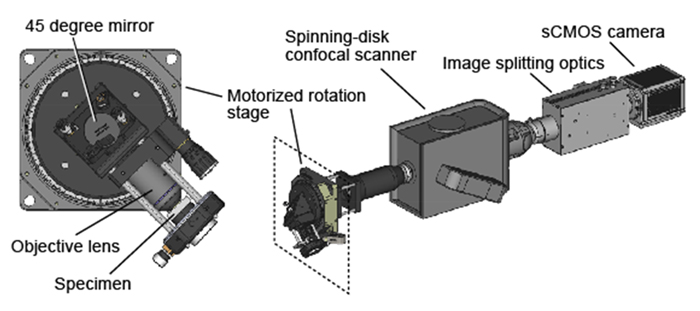
Figure 5. Tiltable objective microscope.
Consistent with the morphological hair-bundle polarity, hair cells medial to the LPR are activated by the lateral-down roll tilts, whereas those lateral to the LPR are activated by the medial-down tilts (Figure 6). In response to the nose-down pitch tilts, hair cells medial to the LPR in the rostral utricle and those lateral to the LPR in the caudal utricle are activated, whereas the rest of the hair cells are activated by the tail-down tilts. Interestingly, hair cells in the medial utricle exhibited larger responses to the head tilt compared to the lateral hair cells. In contrast to the responses to the head tilt, the vibratory stimulus in the pitch or roll axis activated the hair cells only in the rostral and lateral utricle near the LPR.
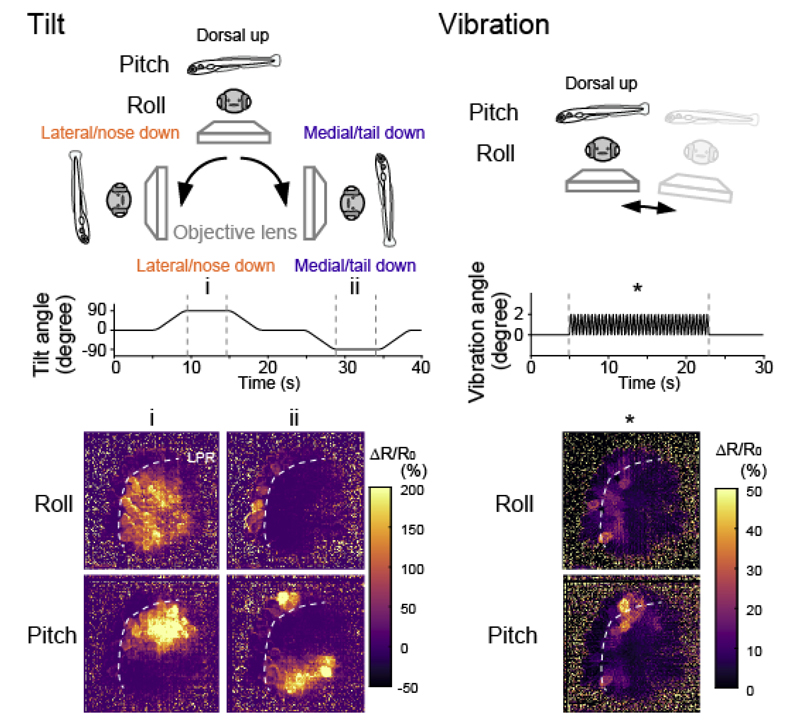
Figure 6. Hair cell responses to head tilt or vibration in the utricle
Together, the tiltable objective microscope visualized, for the first time to our knowledge, the topographically organized response selectivity for the stimulus direction and modality in the vestibular periphery. The imaging strategy we have established here is applicable to the central nervous system, and thus it will provide deeper understanding of the vestibular processing in the brain.












