Basic Biology Program, SOKENDAI (the Graduate University for Advanced Studies)
NIBB Academic Research Experience Program
(formerly known as the NIBB Internship Program)
The NIBB AREP 2025 application has been closed.




About the Academic Research Experience Program (AREP)
formerly known as the NIBB Internship Program
The National Institute for Basic Biology (NIBB) conducts an academic research experience program for undergraduate and graduate students.
NIBB promotes biological sciences by conducting first-rate research to uncover the basic principles common to all life and their diversity. The students at SOKENDAI (the Graduate University for Advanced Studies) receive advanced training at NIBB while doing important research in an excellent research environment. The AREP offers an excellent opportunity to conduct research within ongoing projects at NIBB and participate in educational programs at SOKENDAI.
Research at NIBB encompasses a wide range of biological fields, including cell biology, developmental biology, neurobiology, reproductive biology, evolutionary biology, and environmental biology. Each participant will join an ongoing research project within a world-class research group. Participants can also engage in various academic activities, such as journal clubs and seminars presented by prominent researchers from within and outside NIBB.
The AREP offers an excellent opportunity to experience a part of our Ph.D. program for those considering applying to the Ph.D. course at SOKENDAI. Participants can choose from ten different host laboratories. Please click on link for a complete list.
On-site accommodation is provided at our institute lodge. NIBB covers round-trip international airfare, transportation costs within Japan, and accommodation expenses at the lodge. Participants are responsible for their daily living expenses. Please note that due to budgetary constraints, full reimbursement may not be possible in all cases.
Standard schedule of the AREP:
April to early May: Application period
May to June: Application screening
July to December: Program start*
*The AREP duration will be determined in consultation with your host laboratory and will typically range from several weeks to several months.
For application details, including information about the AREP, please visit link.
For further information about SOKENDAl's Ph.D. program, please refer to the NIBB and SOKENDAI websites.
The AREP schedule is subject to change due to unforeseen circumstances. Participants will be notified directly of any changes.
Application Instructions
Before applying, please review and agree to the terms outlined here.
By submitting your application, you indicate your consent to these terms.
Please email the following three application documents to intern@nibb.ac.jp with
the subject line "The NIBB AREP 2025".
1. Application form
Download the application form here.
Please include a cover letter (approximately 1000 words) explaining why you are a strong candidate for this program. If you are interested in applying to SOKENDAl's Ph.D. program, please state this clearly in your letter.
2. Curriculum Vitae (CV)*
3. Letter of Recommendation*
Please submit one or more letters of recommendation from your principal investigator(s) (PIs) and/or supervisors.
* You may submit your CV and letters of recommendation in any format you prefer.
The AREP 2025 Schedule:
| March 17 (2025) | Application period opens |
| May 6 (Tue) | Application deadline |
| May | Application screening |
| Early-June | Notification of screening results |
| July to December | The AREP implementation period |
Download Applcation Form
Contact
The NIBB Academic Research Experience Program Committee
National Institute for Basic Biology
Basic Biology Program,
The Graduate University for Advanced Studies, SOKENDAI
E-mail: intern@nibb.ac.jp
Host Labs
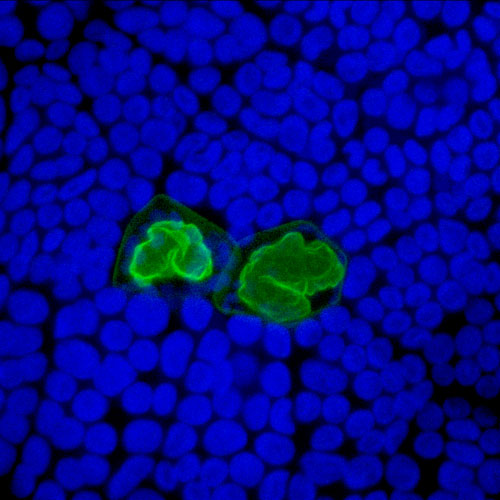
DIVISION OF CELLULAR DYNAMICS
UEDA, Takashi
E-mail :
tueda@nibb.ac.jp
Lab Website
Membrane trafficking among single membrane-bounded organelles plays pivotal roles in various cell activities in eukaryotic cells, which are also critical in multiple layers of higher-ordered functions of multicellular organisms. Although the basic framework of membrane trafficking is well conserved among eukaryotic lineages, recent studies have also suggested that each lineage has acquired a unique membrane trafficking system during evolution. Our research focuses on mechanisms of diversification of membrane trafficking. We are especially interested in how membrane trafficking pathways have been diversified during land plant evolution, which should be also associated with neofunctionalization and/or novel acquisition of organelles in plants. For insights into these points, we are currently conducting comparative analyses between Arabidopsis thaliana and the liverwort, Marchantia polymorpha. Systematic analyses of machinery components of membrane trafficking such as RAB GTPases and SNARE proteins are revealing unique evolutionary paths, which angiosperms and bryophytes followed during evolution.

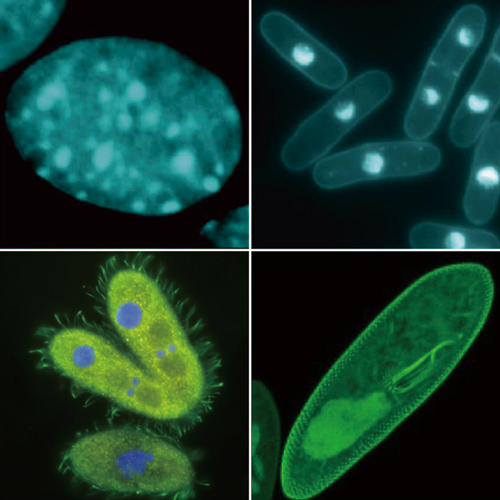
DIVISION OF CHROMATIN REGULATION
Nakayama, Jun-ichi
E-mail :
jnakayam@nibb.ac.jp
Lab Website
Multicellular organisms are made up of diverse populations of many different types of cells, each of which contains an identical set of genetic information coded in its DNA. Cell differentiation and the process of development itself depend on the ability of individual cells to maintain the expression of different genes, and for their progeny to do so through multiple cycles of cell division. In recent years, we have begun to understand that the maintenance of specific patterns of gene expression does not rely on the DNA sequence, but rather takes place in a heritable, “epigenetic” manner. DNA methylation, chromatin modifications, and RNA silencing are some of the best known epigenetic phenomena. Our division investigates how modifications to the structure and configuration of chromatin (complexes of nuclear DNA and proteins) contribute to epigenetic gene regulation by studying events at the molecular scale in the model organism, fission yeast, ciliate Tetrahymena, and in cultured mammalian cells.

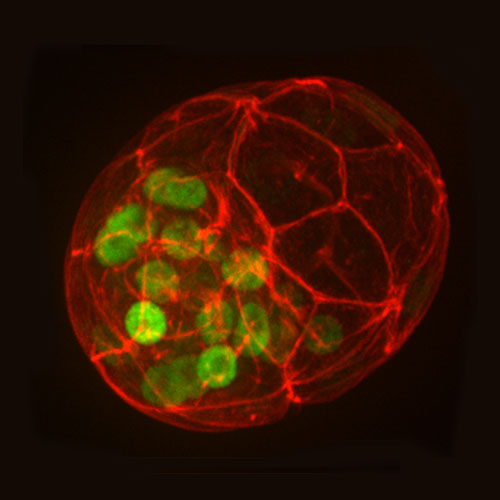
DIVISION OF EMBRYOLOGY
FUJIMORI, Toshihiko
E-mail :
fujimori@nibb.ac.jp
Lab Website
The aim of our research is to understand the events underlying early mammalian development during the period from the pre-implantation to establishment of the body axes. An understanding of early events during embryogenesis in mammals, as compared to other animals, has been relatively delayed. This is mainly due to the difficulties in approaching the developing embryos in the uterus of the mother. The other characteristic of mammalian embryos is their highly regulative potential. The pattern of cell division and allocation of cells within an embryo during the early stages vary between embryos. The timing of the earliest specification events that control the future body axes is still under discussion. Functional proteins or other cellular components have not been found that localize asymmetrically in the fertilized egg. We want to provide basic and fundamental information about the specification of embryonic axes, behaviors of cells and the regulation of body shape in early mammalian development.

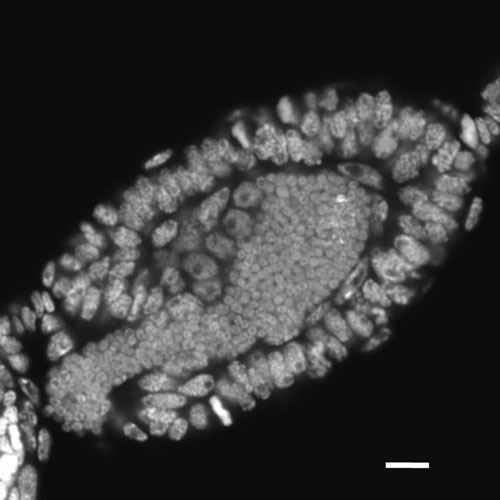
LABORATORY OF EVOLUTIONARY GENOMICS
SHIGENOBU, Shuji
E-mail :
shige@nibb.ac.jp
Lab Website
The term "symbiogenomics” (= symbiosis + genomics) was coined to describe the use of “omic” approaches for defining symbiosis in molecular terms, aiming to understand the network of biological interactions at molecular and genetic levels. We pioneered the symbiogenomics of host-microbe interface in symbioses observed in insects. We mainly study the symbiosis between the pea aphid and the bacterial symbiont Buchnera as a model. Aphid species bear intracellular symbiotic bacteria in the cytoplasm of bacteriocytes, specialized cells for harboring the bacteria. The mutualism is so obligate that neither can reproduce independently. We take advantage of state-of-the-art genomics, such as next-generation sequencing technologies, which has allowed us to find key molecules for symbioses including a transcription factor expressed preferentially in the bacteryocyes and a novel secretion peptide. References: Shigenobu, S., and Wilson, A.C.C. (2011). Genomic revelations of a mutualism: the pea aphid and its obligate bacterial symbiont. Cell. Mol. Life Sci. 68, 1297–1309.

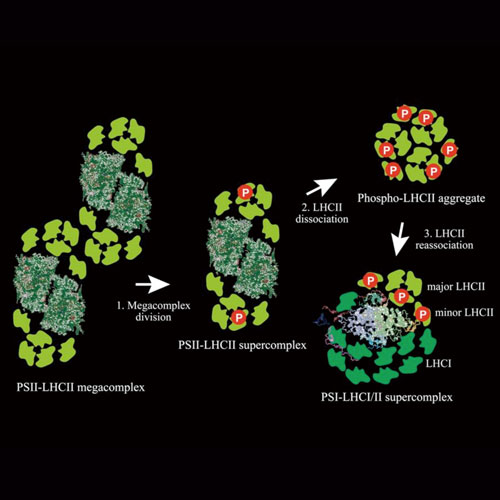
DIVISION OF ENVIRONMENTAL PHOTOBIOLOGY
MINAGAWA, Jun
E-mail :
minagawa@nibb.ac.jp
Lab Website
Plants and algae have a large capacity of acclimating themselves to changing environments. We are interested in these acclimation processes, such as, how efficiently yet safely they harness sunlight for photosynthesis under the changing light environment. Using green algae and plants, we are studying the molecular mechanisms underlying the photoacclimation of the photosynthetic machinery. Our main interests are focused on how the giant chlorophyll-protein supercomplexes, including photosystem I and II, are remodeled or reorganized during photoacclimation events such as state-transitions and non-photochemical quenching, which is explored by applying structural biology, biochemistry, spectroscopy and molecular genetics.

DIVISION OF PLANT ENVIRONMENTAL RESPONSES
MORITA, Miyo T.
E-mail :
mimorita@nibb.ac.jp
Lab Website
Plant organs are capable of sensing various vectorial stimuli, such as light, gravity, touch, and humidity. Plants then reorient their growth direction so as to be in a suitable position to survive and acclimate to their environment. These type of responses of plant organs to directional stimuli are referred to as tropisms. We are conducting research with the greatest focus on the molecular mechanism of gravitropism using Arabidopsis thaliana. In gravity sensing cells, plastids accumulating starch at a high-density (amyloplasts) relocate toward the direction of gravity. Amyloplast relocation serves as a physical signal trigger for biochemical signal transduction, which in turn leads to the regulation of polar auxin transport necessary for change in the direction that a given plant is growing. We are currently investigating how the gravity sensing cell detect the position of amyloplast and how the positional signal is linked to the regulation of polar auxin transport. Gravitropism also affects overall architecture of plants by controlling the growth angles of lateral roots and branches. We also investigate the mechanism underlying the plant architecture controlled by gravity.

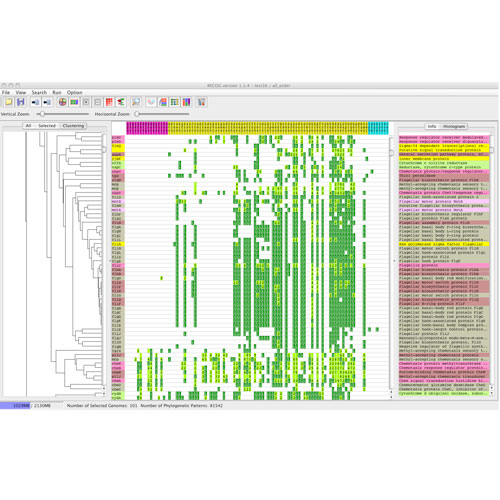
LABORATORY OF GENOME INFORMATICS
UCHIYAMA, Ikuo
E-mail :
uchiyama@nibb.ac.jp
Lab Website
The accumulation of biological data has recently been accelerated by various high-throughput “omics” technologies such as genomics, transcriptomics, proteomics, and so on. The field of genome informatics is aimed at utilizing this data, or finding some principles behind the data, for understanding complex living systems by integrating the data with current biological knowledge using various computational techniques. In this laboratory we focus on developing computational methods and tools for comparative genome analysis, which is a useful approach for finding functional or evolutionary clues to interpreting the genomic information of various species. The current focus of our research is on the comparative analysis of microbial genomes, the number of which has been increasing rapidly. Extracting useful information from such a growing number of genomes is a major challenge in genomics research. Interestingly, many of the completed genomic sequences are closely related to each other. We are now trying to develop methods and tools to conduct comparative analyses not only of distantly related genomes but also of closely related genomes, since we can extract different types of information about biological functions and evolutionary processes from comparisons of genomes at different evolutionary distances.

LABORATORY FOR SPATIOTEMPORAL REGULATIONS
NONAKA, Shigenori
E-mail :
snonaka@nibb.ac.jp
Lab Website
In spite of superficially bilateral symmetry, our bodies are highly asymmetric along the left-right (L-R) axis, for example in the placement of internal organs. Our main aim is to clarify the mechanism by which mammalian embryos generate and establish the L-R asymmetry. Also, we are working for the application of light-sheet microscopy, which enables deep and long-term imaging of living specimens, such as gastrulating mouse embryos.

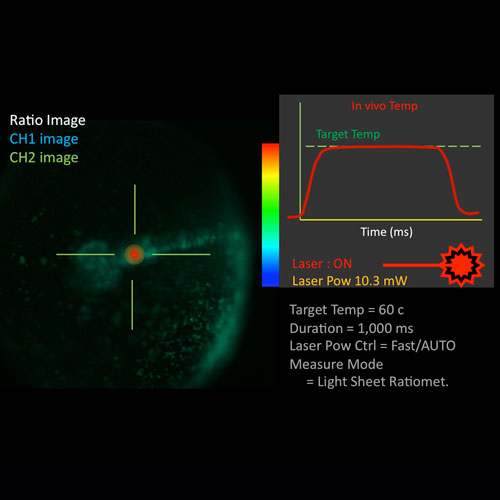
OPTICS AND IMAGING FACILITY
KAMEI, Yasuhiro
E-mail :
ykamei@nibb.ac.jp
Lab Website
Our group is developing gene-expression manipulation methods using ‘light’. Studies of cell fates, cell-cell interaction, and analysis of non-cell autonomous phenomena require an experimental system with fine control of gene expression. To achieve timing-controlled gene expression at the single-cell level we have developed a microscope, IR laser evoked gene operator (IR-LEGO), using the heat shock stress response system (Ref.). This system can be applied to many species, such as C. elegans, zebrafish, medaka and Arabidopsis. Currently we are mainly studying single cell gene knock-out systems using the cre-loxP recombination system applied to medaka mutants. Ref.: Kamei Y., Suzuki M., Watanabe K., et al. (2009) Infrared laser-mediated gene induction in targeted single cells in vivo. Nature Methods 6, 79-81.

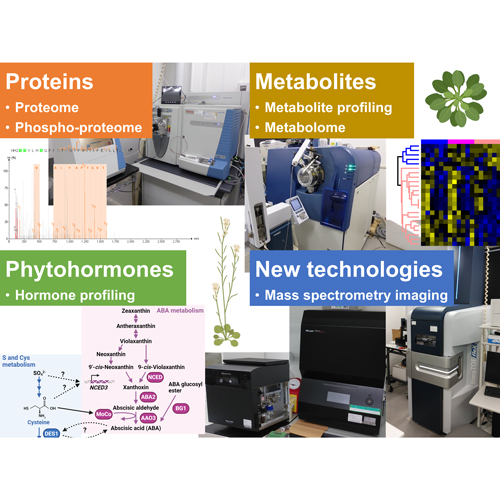
TRANS-OMICS FACILITY
YOSHIDA, Takuya
E-mail :
tyoshida@nibb.ac.jp
Lab Website
Plant growth, development, and stress responses are finely coordinated through the integration of external environmental signals—such as light, temperature, and water availability—with internal signaling pathways. Among these, plant hormones serve as essential endogenous regulators, orchestrating various physiological processes. While the perception and signal transduction mechanisms of plant hormones have been elucidated at the molecular level, and key transcription factors involved in these pathways have been characterized, much remains unknown about the quantitative and qualitative changes in numerous intracellular biomolecules (e.g., proteins, amino acids, and metabolites). To gain a comprehensive understanding of plant hormone responses, our group employs omics-based approaches, including proteomics and metabolomics, to globally analyze proteins and metabolites. Additionally, we are actively developing novel mass spectrometry technologies, which are crucial for advancing these analytical methods.

Contact
The NIBB Academic Research Experience Program Committee
National Institute for Basic Biology
Basic Biology Program,
The Graduate University for Advanced Studies, SOKENDAI
E-mail: intern@nibb.ac.jp