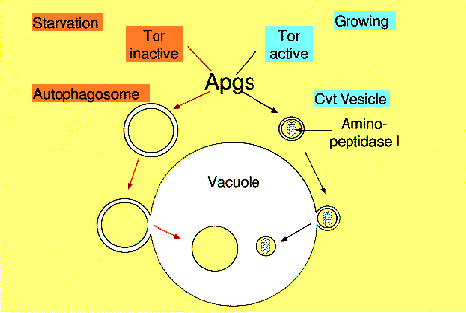
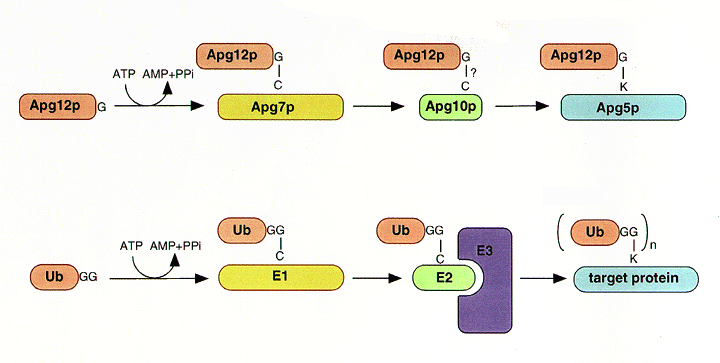
NATIONAL INSITUTE FOR BASIC BIOLOGY

National Institute for Basic Biology
DIVISION OF BIOENERGETICS
- Professor:
- Yoshinori Ohsumi
- Associate Professor:
- Tamotsu Yoshimori
- Research Associate:
- Takeshi Noda
- Yoshiaki Kamada
- Post Doctral Fellow:
- Noboru Mizushima 1
- Takahiro Shintani 2 (from April)
- Naotada Ishihara 1 (from July)
- Takahiro Shintani 2 (from April)
- Graduate Students:
- Takayoshi Kirisako (Graduate Univ. of Advanced Studies)
- Yoshinobu Ichimura (Graduate Univ. of Advanced Studies)
- Technical Staff:
- Yukiko Kabeya
- Visiting Fellow:
- Fumi Yamagata (Teikyo Univ. of Sci. and Tech.)
- Visiting Scientist:
- Kazuaki Furukawa (from November)
- Satsuki Okamoto