16. Available full
length cDNA
The
following eight full length cDNA libraries and full length cDNA clones were
made and will be available from RIKEN BioResource Center (http://www.brc.riken.jp/lab/epd/Eng/).
Strain: Physcomitrella
patens (Hedw.)
Bruch and Schimp subsp. patens
We
used an original strain instead of Gransden2004 used for whole genome shotgun
sequence by JGI and ESTs may include SNPs.
Distribution: Full length
cDNA clones of pph, pphb, and pphn libraries are distributed from RIKEN
BioResourceCenter (http://www.brc.riken.jp/lab/epd/Eng/species/moss.shtml).
Clones of other libraries will be distributed from the same center after
publication of the corresponding paper. Before the publication, those clones
will be shared as a collaboration work and contact to Dr. Mitsuyasu Hasebe
(mhasebe [at mark] nibb.ac.jp).
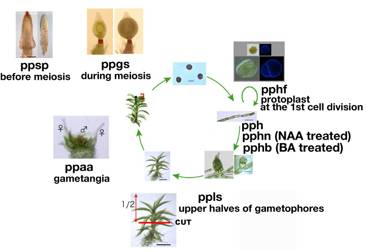
(1)
pph (Nishiyama et al. 2003 PNAS 100: 8007-8012)
Protonemata
and young gametophore without exogenous phytohormones
The tissue from which RNA was extracted
contained chloronemata and young gametophores with 2 to 5 leaves.
Protonemata were blended by the POLYTRON,
and then cultivated on the BCDATG medium for 13-14 days under the continuous
light.
Full length
cDNA library. A backbone of the vector is basically from pBluescript(KS),·@
that was in vivo excised from a modified lPS phage vector (Mo bi Tec,
Germany). 5' end of the cDNA that was digested with XhoI was ligated to SalI
site of the vector and the 3' end including polyA tail was ligated to BamHI
site of the vector.·@ cDNA instert could be amplified with conventional
T7 and T3 primers.·@ This full-length
cDNA library was generated basically according to the method described in
Seki M. et al. (The Plant J 15, 707-720 (1998)).
(2)
pphb (Nishiyama et al. 2003 PNAS 100: 8007-8012)
Protonemata treated with
benzylaminopurine
Protonemata were blended by the POLYTRON,
and then cultivated on the BCD medium containing 0.5uM BA (benzylaminopurine)
for 8 to 13 days under the continuous light.·@
The tissue from which RNA was extracted
contained chloronemata, caulonemata and malformed buds.
Normalized
full length cDNA library. A backbone of the vector is pBluescript II, that
was in vivo excised from a modified lPS phage vector (Mo bi Tec, Germany).
XhoI digested-5' end of cDNA is ligated to SalI site of the vector, and the
BamHI digested-3' end including poly-A tail is ligated to BamHI site of the
vector. cDNA instert could be amplified with conventional T7 and T3 primers.
This normarized full-length cDNA library was generated basically according
to the method described in Genome Research 10, 1617-1630 (2000), Carninci,
P. et al.
(3)
pphn (Nishiyama et al. 2003 PNAS 100: 8007-8012)
Protonemata treated with NAA
Protonemata were blended by the POLYTRON,
and then cultivated on the BCD medium containing 1uM NAA (naphthalene acetic
acid) for 8 to 11 days under the continuous light.·@
The tissue from which RNA was extracted
contained chloronemata, caulonemata and rhizoid-like protonemata.
Normalized
full length cDNA library. A backbone of the vector is pBluescript II, that was
in vivo excised from a modified lPS phage vector (Mo bi Tec, Germany). XhoI
digested-5' end of cDNA is ligated to SalI site of the vector, and the BamHI
digested-3' end including poly-A tail is ligated to BamHI site of the vector.·@ cDNA instert could be amplified with
conventional T7 and T3 primers. This normarized full-length cDNA library was
generated basically according to the method described in Genome Research 10,
1617-1630 (2000), Carninci, P. et al.
(4)
pphf (Nishiyama et al. submitted)
Protoplasts at the first cell division
Physcomitrella patens protonemata
were inoculated on BCDATG medium for every ca. 5 days.·@ Protoplasts were isolated from the
protonemata, further incubated at 25C under continuous light for 2-3 days.·@ The regenerated cells, which were rich in
cells at the stage during the first asymmetric cell division, were
collected.·@ Total RNA was extracted for
constructing a full-length cDNA library.
cDNA library was prepared by the
vector-capping method (Kato et al. 2005. DNA Res. 12: 53-62).
vector
information
(5)
ppsp: Sporophytes
before meiosis
The gametophores harboring gametangia were
further cultivated at 15·┴C under the short day conditions for 21 to 28 days to
collect sporophyte tissue before meiosis. Archegonial tissue was included in
the sample.
cDNA
library was prepared by the oligo-capping method (Maruyama and Sugano 1994.
Gene 138: 171-174).
(6)
ppls: Upper
halves of gametophores (Nishiyama
et al. submitted)
Gametophores were cultivated on sterile
peat pellets (Jiffy-7; Jiffy Products International AS, Kristansand, Norway)
for 1 to 1.5 months at 25·┴C under continuous light.
cDNA library was prepared by the
oligo-capping method (Maruyama and Sugano 1994. Gene 138: 171-174).
(7)
ppaa: Gametophore
tips including the antheridia and archegonia (Nishiyama
et al. submitted)
Gametophores cultivated under the same
conditions for the same period were moved to 15·┴C under 8-h light/16-h dark
conditions to induce gametangia and cultivated for 11 to 26 days. Those
archegonia that were brown in color, which is an indicator of fertilization,
were discarded.
cDNA
library was prepared by the oligo-capping method (Maruyama and Sugano 1994.
Gene 138: 171-174).
(8) ppgs: Sporophytes during meiosis
The
gametophores harbouring gametangia were further cultivated at 15·┴C under the
short day conditions for 31 to 53 days to collect sporophyte tissue during
meiosis. Archegonial tissue was removed and only sporophytic tissue was
collected.
cDNA library
was prepared by the oligo-capping method (Maruyama and Sugano 1994. Gene 138:
171-174).