| DIVISION OF BIOLOGICAL REGULATION AND PHOTOBIOLOGY (ADJUNCT) | ||
|
||
Plants respond to light as an environmental
factor to optimize development and regulate other physiological phenomena.
Phytochrome and blue light receptors, such as cryptochrome and phototropin
(phot), are the main photoreceptors for plant photomorphogenesis.
The goal of our research is to elucidate the photoperception and the
signal transduction pathways of photo-morphogenesis. One of our major subjects is chloroplast photo-relocation movement,
which is thought to be one of the simplest phenomena in this field.
We use the fern Adiantum capillus-veneris and the moss Physcomitrella
patens as model plants for our cell biological approach since
the gametophytes are very sensitive to light and the organization
of the cells is very simple. We also use Arabidopsis mutants
to identify the genes regulating chloroplast photo-relocation movement. Chloroplasts accumulate at the cell surface under weak light and escape
from the cell surface to the anticlinal wall under strong light to
optimize photosynthesis. We identified the photoreceptors in Arabidopsis.
Phototropin2 (phot2) mediates the avoidance response under strong
light and phot1 and phot2 mediate, redundantly, the accumulation response
under weak light. However, components of signal transduction pathways
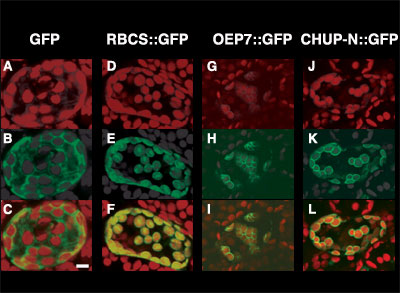
still remain to be identified. A mutant called chup1 is deficient
in chloroplast movement and consequently chloroplasts gather at the
bottom of cells. CHUP1 is a novel gene that has a hydrophobic
region in the N-terminus and an actin binding domain, a prolin-rich
region, and two leucin zippers. It was confirmed that the actin-binding
domain binds F-actin. When fusion proteins of the N-terminal, hydrophobic
region of CHUP1 with GFP were expressed transiently in chup1
mutant cells, fluorescence of the fusion proteins were found in the
outer part of chloroplasts, likely on the outer membrane of chloroplasts.
CHUP1 is proposed to play an important role on chloroplasts movement
of both accumulation and avoidance responses. Adiantum phytochrome3 (PHY3) is a unique chimeric protein
with phytochrome structure in the N-terminal half and phototropin
structure in the C-terminal half. Transient expression of PHY3
or modified PHY3 genes in rap (red light-induced
aphototropic) mutants of Adiantum reveal that phy3 is the
photoreceptor of red light-induced chloroplast photorelocation movement,
and that the phy3 signal might be transferred through the C-terminal,
phototropin region. The phot region of phy3 has not yet been shown
to function as a blue light photoreceptor, but the above method of
transient expression of modified PHY3 in rap and
phot mutants may provide experimental evidence of this possibility
soon. In order to elucidate the role of genes in Adiantum and rice,
we have tried to establish new methods for gene targeting in these
organisms. Transposable elements constitute a large portion of eukaryotic genomes and contribute to their evolution and diversification. Miniature inverted-repeat transposable elements (MITE) constitute one of the main groups of transposable elements. MITEs have been found in wide range of organisms but active MITEs have not been identified. We found a new class of MITEs in rice and named them miniature Ping (mPing). mPing was identified as the first active MITE from any organism and the first active DNA transposon from rice. mPing is a short 430 base pairs element with 15 base pair terminal inverted repeats that lacks a transposase. mPing elements are activated in calli derived from anther culture and excise efficiently from original sites to reinsert into new loci. The mPing-associated Ping element which has a putative transposase sequence was also found and shown to transpose within the rice genome. Rice is the most agriculturally important crop in the world. mPing/Ping transposon system is a useful molecular tool for gene isolation and
gene knockout in rice. To understand the genetic information of a fern, Adiantum capillus-veneris,
a normalized cDNA library was constructed from prothallia grown under
white light and analysis of expressed sequence tags (EST) were carried
out. Approximately 10,000 clones were sequenced and clustering of
these obtained sequences was performed. As a result, 7,132 non-redundant
groups were generated. These groups were subjected to similarity searches
to identify putative function. Approximately 1,600 EST groups were
found to be similar to sequences of genes registered in the public
database. About 1,100 EST groups showed similarity to sequences of
unknown function. The remaining EST groups showed no significant similarity
and were classified as novel sequences.
|
||
| Publication List: (1) Original articles Stoelzle, S., Kagawa, T., Wada, M., Hedrich, R., Dietrich, P. (2003) Blue light activates calcium-permeable channels in Arabidopsis mesophyll cells via the phototropin signaling pathway. Proc. Natl. Acad. Sci. USA 100, 1456-1461. Sato, Y., Wada, M., Kadota, A. (2003) Accumulation response of chloroplasts induced by mechanical stimulation in bryophyte cells. Planta 216, 772-777. Iwata, T., Nozaki, D., Tokutomi, S., Kagawa, T., Wada, M., Kandori, H. (2003) Light-induced structural changes in the LOV2 domain of Adiantum phytochrome3 studied by low-temperature FTIR and UV-visible spectroscopy. Biochemistry 42, 8183-8191. Oikawa, K., Kasahara, M., Kiyosue, T., Kagawa, T., Suetsugu, N., Takahashi, F., Kanegae, T., Niwa, Y., Kadota, A., Wada, M. (2003) CHUP1 is essential for proper chloroplast positioning. Plant Cell 15, 2805-2815. Srinivas, A., Behera, R. K., Kagawa, T., Wada, M., Sharma, R. (2003) High pigment1 mutation negatively regulates phototropic signal transduction in tomato seedlings. Plant Physiol. in press Lamparter, T., Kagawa, T., Brücker, G., Wada, M. (2003) Positive and negative tropic curvature induced by microbeam irradiation of protonemal tip cells of the moss Ceratodon purpureus. Plant Biology in press. Review articles Sato, Y., Kadota, A., Wada, M. (2003) Chloroplast movement: dissection of events downstream of photo- and mechano-perception. J. Plant Res. 116, 1-5. Wada, M., Kagawa, T., Sato, Y. (2003) Chloroplast movement. Annu. Rev. Plant Biol. 54, 455-468. Wada, M. (2003) Blue light receptors in fern and moss. In: ESP Comprehensive Series in Photoscience, Photoreceptors and Light Signaling. Ed. By Batchauer, Elsevier Science Publishers. Kasahara, M., Wada, M. (2004) Chloroplast avoidance movement. In: Annual Plant Reviews “Plastids” Ed. by Möller, Kruwer Academic Publishers, in press. Wada, M., Kanegae, T. (2004) Photomorphogenesis of Ferns. In: Photomorphogenesis in Plants 3ed Edition. Ed. by Schäfer and Nagy, Kruwer Academic Publishers, in press. |