National Insitute for Basic Biology

National Insitute for Basic Biology

|
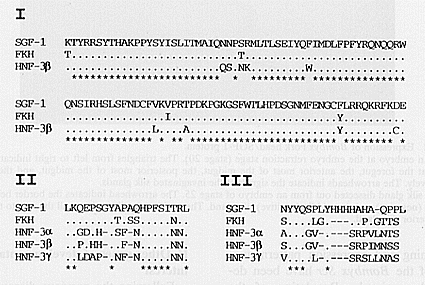
| Fig.1 Comparison of SGF-1 with other members of Fork head/HNF-3 family. The three domains conserved between Drosophila Fork head and mammalian HNF-3 factors are present also in SGF-1. The domain boundaries in SGF-1 are Lys103/Glu212(I), Leu286/Leu303(II), and Asp331/Leu349(III). Domain I is the DNA binding domain, and domains II and III participate in transactivation by HNF-3 beta. |
|
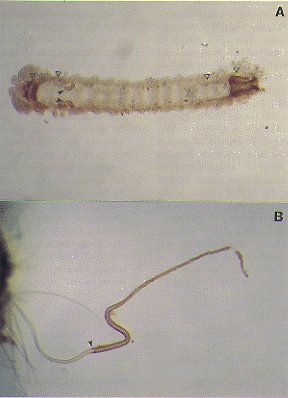
| Fig.2 Expression of Bombyx Fork head/SGF protein. A. An embryo at the embryo retraction stage (stage 20). The triangles from left to right indicate positive signals at the foregut, the anterior most of the midgut, the posterior most of the midgut, and the hindgut, respectively. The arrowheads indicate the signals at the invaginated silk glands. B. A silk gland dissected out from an embryo of stage 25. The arrowhead indicates the border between the anterior (negative) and the middle (positive) silk gland. The positive signal continues all the way to the end of the posterior silk gland. |