11.6
Overexpression
Minoru Kubo, Tomomichi
Fujita, and Mitsuyasu Hasebe
Introduction
Gain-of-function
experiments using over expression and ectopic expression are indispensable to
analyze gene function. While the cauliflower mosaic virus (CaMV)
35S promoter is a strong promoter in flowering plants, in Physcomitrella patens, CaMV35S promoter is strong enough to induce
antibiotic resistance gene for selection but is too weak to see the effects of
over expression and ectopic expression for genes involved in development. For
stronger induction, the rice actin promoter (Zhang et al. 1991), the 7113
promoter modified from CaMV35S (Mitsuyara et al.
1996), and the PpEF1-¿
promoter (Kubo et al unpublished) are useful. The actin promoter works well in
protonemata but is much weaker in gametophores. On the other hand, the 7113 and
PpEF1-¿promoters
work in almost tissue including gametophores, although it seems that not all of
the cells in gametophores show overexpression of genes. In this way, you have
to select the promoter for overexpression in accordance with your purpose.
To introduce the construct of
overexpression to P. patens genome,
some targeting sites, the PpMADS2, Pphb7, BS213 have
been reported (Pphb7, Sakakibara et al. 2003; BS213, Schaefer et
al. 1997). Additionally, we established new targeting sites, PIG1 and PTA1,
by using informatics and genome resource of P.
patens (Kubo et al. unpublished).
Here we display the list and schematic representations of available vectors for
overexpression using various promoters and targeting sites.
Materials
and Methods
Construction
of a targeting vector
Conventional
methods
The
PCR fragment with blunt ends, which is amplified with proof-reading DNA
polymerase such as KOD plus DNA polymerase (TOYOBO), can be directly cloned in
the EcoRV
and SmaI
sites of these vectors.
Gateway
compatible
Your
gene and DNA fragment, which is amplified with proof-reading DNA polymerase
such as KOD plus DNA polymerase (TOYOBO), are subcloned
into pENTR/D/TOPO vector. Your gene or DNA fragments
in these pENTR/D/TOPO vectors (entry clones) are
integrated into destination vectors by the LR reaction. For details of the
GATEWAY system (Invitrogen), you can refer to websites as follows: http://www.invitrogen.com/site/us/en/home/Products-and-Services/Applications/Cloning/Gateway-Cloning.html
Transformation
and selection of candidate lines
See Chapter 11.3
Overexpression
vector list
|
vector |
promoter |
targeting site |
selection |
Vector map |
|
pPpMADS2 7113 |
7113 |
PpMADS2 |
G418 |
(1) |
|
pPpMADS2 7113 rev |
7113 |
PpMADS2 |
G418 |
(1) |
|
pPphb7 7113 |
7113 |
Pphb7 |
G418 |
(1) |
|
pPphb7 7113 rev |
7113 |
Pphb7 |
G418 |
(1) |
|
pPpMADS2 Actin |
ACT |
PpMADS2 |
G418 |
(2) |
|
pPpMADS2 Actin rev |
ACT |
PpMADS2 |
G418 |
(2) |
|
pTFH15.3 |
ACT |
Pphb7 |
G418 |
(3) |
|
pBS213-7113-mRFP-G |
7113 |
BS213 |
Zeo |
(4) |
|
pBS213-7113-G-mRFP |
7113 |
BS213 |
Zeo |
(4) |
|
pBS213-7113-citrine-G |
7113 |
BS213 |
Zeo |
(4) |
|
pBS213-7113-G-citrine |
7113 |
BS213 |
Zeo |
(4) |
|
pBS213-7113-HA-G |
7113 |
BS213 |
Zeo |
(4) |
|
pBS213-7113-G-HA |
7113 |
BS213 |
Zeo |
(4) |
|
pPOG1 |
PpEF1-¿ |
PIG1 |
Hyg |
(5) |
|
pPOYG1 |
PpEF1-¿ |
PIG1 |
Hyg |
(5) |
|
pPOCG1 |
PpEF1-¿ |
PIG1 |
Hyg |
(5) |
|
pT1OG |
PpEF1-¿ |
PTA1 |
Zeo |
(6) |
|
pT1OGY |
PpEF1-¿ |
PTA1 |
Zeo |
(6) |
|
pT1OGC |
PpEF1-¿ |
PTA1 |
Zeo |
(6) |
ACT: rice actin promoter, Zeo: zeocin, Hyg:
hygromycin
Remarks
1.
By Pphb7 disruption, its transformants show abnormal development of rhizoids.
Development of other tissue such as protonemata is not distinguishable from
that in wild type (Sakakibara et al. 2003). We have not found any phenotypic change in the
transformants introducing the harmless transgene to PpMADS2, BS213, PIG1 and
PTA1.
2.
Gene silencing is sometimes observed in
some lines with overexpression constructs. The gene silencing is hereditable,
although gene expression is occasionally
recovered in the mucilage hairs of gametophores and the foot of sporophytes.
You should select the transformants with a single insertion, because multi copy
insertion lines have a greater tendency to induce gene silencing than single
ones. To confirm whether the gene is effectively overexpressed, expression
analyses of the transcript and protein are recommended. It might be useful for
monitoring gene expression to construct GFP-fusion protein which maintains
native function in P. patens.
3.
The 7113 promoter was kindly provided by
Dr. I. Mitushara and Dr. Ohashi
(Mitsuhara et al. (1996), Plant Cell Physiol. 37:
49-59). The actin promoter from
rice was kindly provided by Dr. Wu (Zhang et al. (1991), Plant Cell,
3:1155-1165). The rice actin promoter was >10 times stronger than 7113
promoter in P. patens protoplasts
(Fujita et al. unpublished). The
PpEF1-¿promoter
was isolated from P. patens
genome (Kubo et al. unpublished).
Vector map
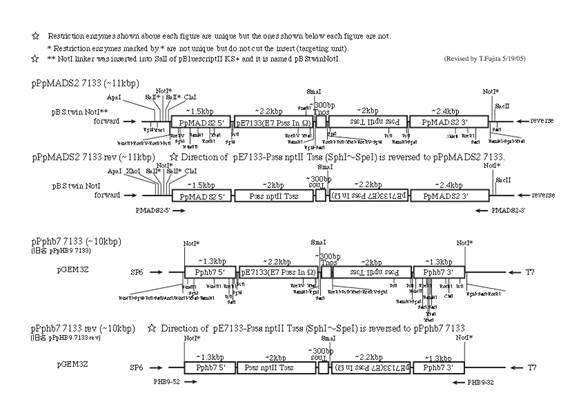
(1)
PpMADS2 target, 7113 promoter
Note: Please substitute gp7113h for
gpE7133h in the figures.

-
E. coli harboring a plasmid with the 7113
promoter should be cultured at 30ºC rather than 37ºC. Truncation of the 7113
promoter sequence is sometimes found at 37ºC.
-
NotI is
usually used to linearize the plasmid for transformation.
(2)
PpMADS2 target, rice actin promoter
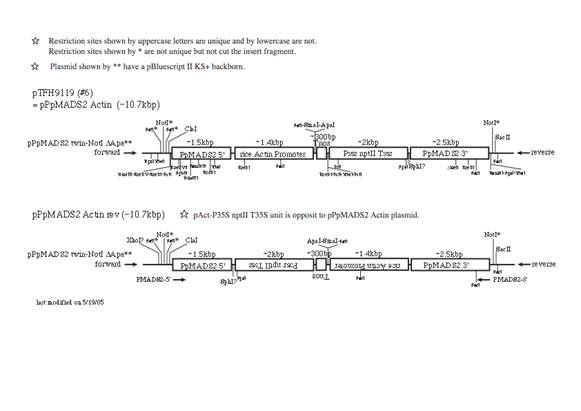
NotI
is usually used to linearize the plasmid for transformation.
(3)
Pphb7 target, rice actin promoter
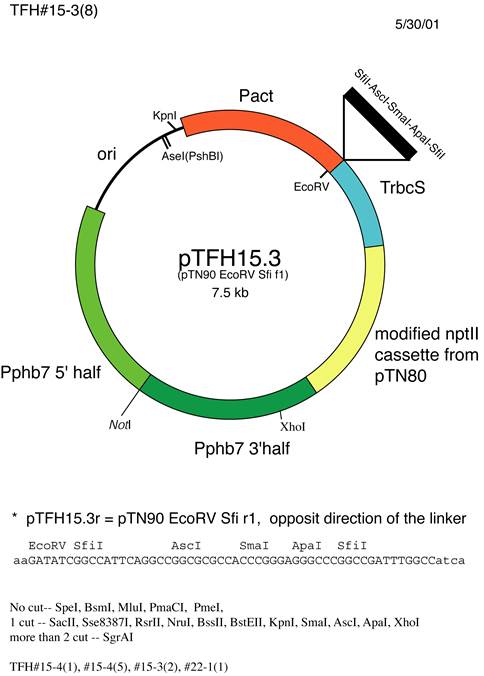
NotI
is usually used to linearize the plasmid for transformation.
(4) BS213 target, 7113 promoter,
gateway compatible, with tag sequence (His, mRFP,
citrine)
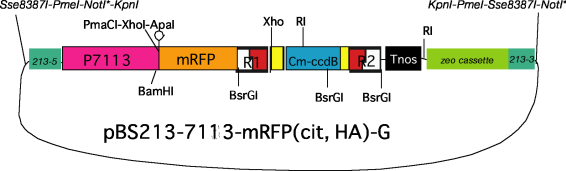
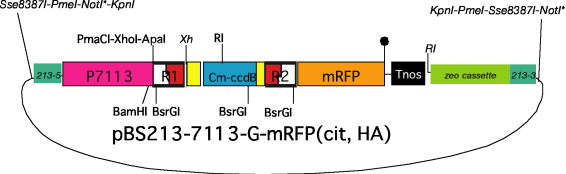
*As
the LR reaction with pENTR Directional TOPO brings a NotI site into
the resultant product, NotI
cannot be used for excision of a
target fragment.
(5)
PIG1 target, PpEF1-¿ promoter, gateway compatible, with tag sequence (citrine
as YFP), cerulean as CFP)
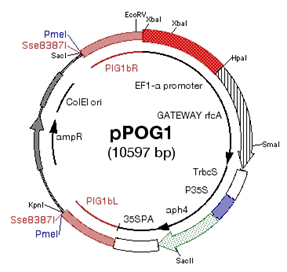
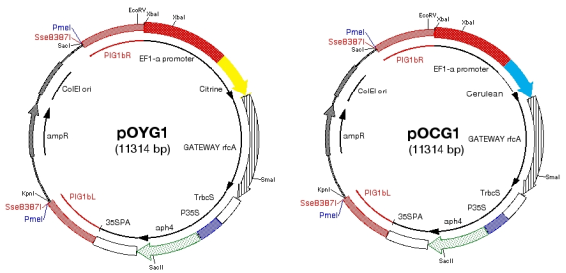
-
PmeI and Sse8387I are usually used to linearize the
plasmid for transformation.
(6) PTA1 target, PpEF1-¿
promoter, gateway compatible, with tag sequence (citrine as YFP), cerulean as
CFP)
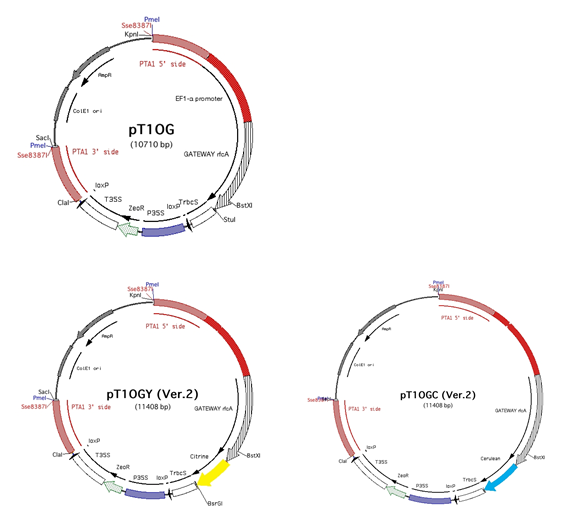
-
PmeI and Sse8387I are usually used to linearize the
plasmid for transformation.