11. Loss/Gain of
Function Analyses
11.1 Gene deletion
Yuji Hiwatashi, Tomoaki Nishiyama
and Minroru Kubo
1. Strategy
This chapter provides a brief outline for gene
deletion by the gene targeting technique. A coding region of a targeted gene
should be fully deleted from its genomic locus to generate a null mutant. A
mere insertion of a selection marker in a coding region may cause expression of
a truncated gene. Polyadenylation signals of the CaMV 35S and the NOS
terminators in the marker cassettes do not completely terminate transcription,
and "read through" from the marker cassette happens. When a specific
domain of a gene is crucial for its function, complete deletion of this domain
may be sufficient for loss of its function.
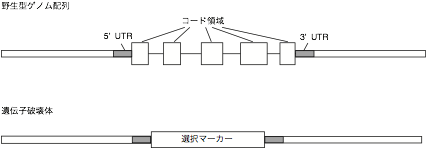
To fully delete a coding region,
genomic DNA fragments 5' and 3' to the coding region are required for
construction of a targeting vector. The genomic DNA fragment is recommended to
be longer than 1 kb. The corresponding genomic fragments are isolated by TAIL-PCR
or by a regular PCR using primers based on genomic sequence information in the DNA
database (e.g. PHYSCObase: http://moss.nibb.ac.jp or the JGI genome
browser: http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html).
2. Construction of a targeting vector
The genomic fragments
corresponding to homologous regions are inserted outside the 5’ and 3’ ends of
a selection marker cassette, respectively. The pTN182, pTN186, p35S-loxP-Zeo,
and p35S-loxP-BSD vectors confer resistance to G418, hygromycin, zeocin, and
blasticidin S, respectively. The PCR fragment with blunt ends, which is
amplified with proof-reading DNA polymerase such as KOD plus DNA polymerase
(TOYOBO), can be directly cloned in the EcoRV
and SmaI sites of pTN182, pTN186, and
p35S-loxP-BSD vectors.
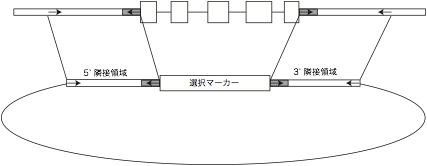
Design
primers to flanking genomic regions for later PCR screening
of transformants. Most right and left primers will be arranged for transformant
screening as in the following figure.

3.
Transformation and selection of candidate lines
The DNA fragment constructed for
targeting is excised from the vector with appropriate restriction enzymes.
Purification of the fragment is not necessary, and we usually use a mixture of
the DNA fragment and a vector for transformation. [Different opinion by Dr.
Andrew Cumming; We think purification of the fragment is necessary!! Actually,
we usually use PCR-amplified fragments.] Approximately 10 – 20 µg of the DNA fragment-vector mixture
is introduced to protoplasts. Lower and higher DNA amount cause less
transformants and more insertions, respectively. Approximately 5 – 10 µg
PCR amplified DNA fragment can also be used for transformation [Comment by Dr.
Andrew Cuming; We usually use 15 micrograms. Less than this and the number of
transformants is reduced].
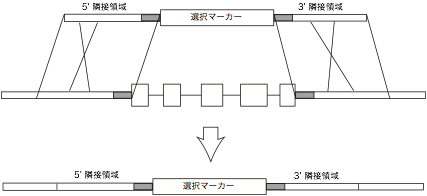
PCR and
Southern blot analyses are used to select transformed lines with a correct
replacement. PCR is used at the first stage of screening, and then southern
analysis is performed. To avoid an unexpected recombination, you definitely
need to perform southern hybridization before proceeding to further analyses of
the transformants.
(Three
kinds of PCR reactions are recommended) see figure below.
(1) Use
primers that anneal to 5' flanking genomic region and to the 5’ side of a
selection marker cassette.
(2) Use
primers that anneal to 3' flanking genomic region and to the 3’ side of a
selection marker cassette.
(3) Use
primers located in the selection marker cassette to eliminate a line with
tandem insertions.



![]()

![]()