
 |
DIVISION OF PHOTOBIOLOGY(ADJUNCT) |
| Professor (Adjunct): | WADA, Masamitsu |
| Associate Professor (Adjunct): | YAMAUCHI, Daisuke |
| Research Associate: | KIKUCHI, Kazuhiro |
| NIBB Research Fellow: | OGURA, Yasunobu |
| MEXT Postdoctoral Fellow: | TAKAHASHI, Fumio |
| TMU Postdoctoral Fellow: | CHRISTENSEN, Steen |
| Visiting Scientists: | OIKAWA, Kazusato |
| SUETSUGU, Noriyuki | |
| UENAKA, Hidetoshi |
|
Plants respond to light as an environmental factor to optimize development and regulate other physiological phenomena. Phytochrome (phy) and blue light receptors, such as cryptochrome (cry) and phototropin (phot), are the main photoreceptors for plant photomorphogenesis. The goal of our research is to elucidate the photoperception and signal transduction pathways of photo morphogenesis. |
I. Chloroplast relocation movementOne of our major subjects is chloroplast photo-relocation movement, which is thought to be one of the simplest phenomena in this field. We use the fern Adiantum capillus-veneris and the moss Physcomitrella patens as model plants for our cell biological approach since the gametophytes are very sensitive to light and the organization of the cells is very simple. We also use Arabidopsis mutants to identify the genes regulating chloroplast photo-relocation movement. |
1-1Arabidopsisn Arabidopsis leaves, chloroplast movement is fluence rate dependent. Under lower light fluence rate, chloroplasts accumulate at the cell surface to maximize photosynthetic potential. Under high fluence rate, chloroplasts avoid incident light to escape photodamage. We examined the phenomenon of chloroplast avoidance movement and demonstrated a proportional relationship between fluence rate and the velocity of chloroplast avoidance movement. When a small area is irradiated with a microbeam, the majority of chloroplasts inside the beam begin moving towards the outside of the beam. Some chloroplasts, however, begin to move only after a relatively long lag time. The length of the lag period becomes longer under higher fluence rate light and occurred more frequently with chloroplasts located nearest the center of the microbeam.In addition, we showed that the amount of light-activated phot2, the photoreceptor for the avoidance response, likely plays a role in this phenomenon, as heterozygous mutant plants show a reduced avoidance velocity compared to that of homozygous wild type plants. |
1-2PhyscomitrellaPhototropin is the blue light receptor that mediates blue and red light induce chloroplast movement in the moss Physcomitrella patens, which has four phototropin genes, PHOTA1, PHOTA2, PHOTB1 and PHOTB2. These genes were classified into two groups (PHOTA and PHOTB) on the basis of their deduced amino acid sequences. Then phototropin disruptants were generated by homologous recombination and used for analysis of chloroplast movement. It was found that blue light-induced chloroplast movement was mediated by phototropins. Both photA and photB groups were able to mediate chloroplast avoidance, although the photA group contributed more to the response. Red light-induced chloroplast movement was also significantly affected in photA2photB1photB2 triple disruptants. Because the primary photoreceptor for red light-induced chloroplast movement in P. patensis phytochromes, phototropins may be downstream components of phytochromes in the signaling pathway. The involvement of phototropins in the phytochrome signaling pathway was reported for the first time. |
1-3AdiantumThe avoidance movement response in Adiantum phot2 deficient mutants can be restored by transient expression of non-mutant AcPHOT2 cDNA, indicating that chloroplast avoidance movement in this fern is mediated by the Acphot2 protein. Further functional analyses of the Acphot2 protein were performed using this transient assay for chloroplast avoidance movement. The results suggest that the LOV2, but not the LOV1, domain of Acphot2 is essential for avoidance movement. |
II. Gene targeting and gene silencingIn order to elucidate the role of genes in Adiantum and rice, we have tried to establish new methods for gene targeting in these organisms. |
2-1Miniature transposable elementTransposable elements constitute a large portion of eukaryotic genomes and contribute to their evolution and diversification. We identified active transposable elements, miniaturePing (mPing), Ping and Pong in rice. The mPing element was identified as the first active MITE from any organism. mPing is a short 430 base pair element with 15 base pair terminal inverted repeats that lacks a transposase. mPingelements are activated in calli derived from anther culture and excise efficiently from original sites to reinsert into new loci. Ping and Pong transposable elements were isolated as putative autonomous elements encoding an IS/PIF/Harbinger superfamily of transposases. Evolutionally, the number of copies of mPing elements has increased in japonica cultivars, but not in indica cultivars and their ancestral species, Oryza rufipogon. Japonica cultivars are the only rice varieties in which the transposable Pingelement can be detected. The mPing/Ping transposon system may prove a useful molecular tool for gene isolation and gene knockout in rice, the most agriculturally important crop in the world. |
2-2DNA interference inAdiantumSilencing of gene expression by RNA interference (RNAi) is a useful technique for determining the roles of genes of unknown function in a wide range of organisms. The dramatic increase in the number of genes of unknown function as a result of whole genome sequencing projects, as well as EST projects, means that faster approaches are needed for understanding gene function. We found a simple method for silencing genes using DNA fragments homologous to the target gene rather than RNAi. We have named this approach DNA interference or DNAi. It has the advantage of being faster and simpler than current RNAi approaches, and allows simultaneous silencing of multiple genes. |
|
|
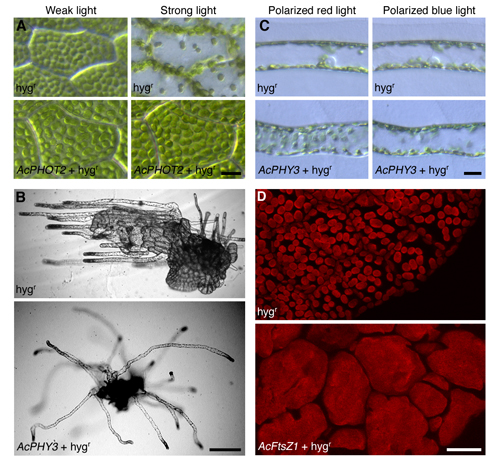
Fig. 1Phenotypes by DNAi. (A) Chloroplast accumulation and avoidance movement in prothalli induced by weak white light (left) and strong blue light (right), respectively. Note that the avoidance response under strong light was inhibited by AcPHOT2 introduction. (B) Red light-induced phototropism in protonemal cells generated from a prothallus.Red light was irradiated from the left. Note that phototropic response was inhibited by AcPHY3 introduction. (C) Chloroplast relocation induced by polarized red (left) or blue light (right) in protonemal cells of (B). Note that chloroplast accumulation response induced by red light was inhibited by AcPHY3 introduction. (D) Chloroplasts of a prothallus. Note that chloroplast division was inhibited by AcFtsz introduction.Both promoterless AcPHOT2 (A), AcPHY3 (B,C), and AcFtsZ1(D)and hygr gene or hygr gene only were introduced into these gametophytes. Bar in A, C and D: 20 µm, B: 200 µm. See Kawai-Tokuoka et al (2004) in detail. |
List of publication: |
Original articles |
|
Srinivas, A., RK Behera, T. Kagawa, M. Wada, and R. Sharma (2004) High pigment1mutation negatively regulates phototropicsignal transduction in tomato seedlings. Plant Physiol.134:790-800. |
|
Lamparter, T., T. Kagawa, G. Brucker and M. Wada (2004) Positive and negative tropic curvature induced by microbeam irradiation of protonemal tip cells of the moss Ceratodon purpureusPlant Biology 6:165-170. |
|
Kagawa, T., M. Kasahara, T. Abe, S. Yoshida and M.Wada (2004) Function analysis of Acphot2 using mutants deficient in blue light-induced chloroplast avoidance movement of the fern Adiantum capillus-veneris L. Plant Cell Physiol. 45: 416-426. |
|
Kagawa, T. and M. Wada (2004) Chloroplast avoidance movement rate is fluence dependent. Photochem. Photobiol. Sci. 3:592-595. |
|
Kasahara, M., T. Kagawa, Y. Sato, T. Kiyosue and M. Wada (2004) Phototropins mediate blue and red light-induced chloroplast movements in Physcomitrella patensPlant Physiology135:1388-1397. |
|
Mochizuki, T., Y. Onda, E. Fujiwara, M. Wada and Y. Toyoshima (2004) Two independent light signals cooperate in the activation of the plastid psbD blue light-responsive promoter in Arabidopsis.FEBS Letters 571:26-30. |
|
Kawai-Toyooka, H., C. Kuramoto, K. Orui, K. Motoyama, K. Kikuchi, T. Kanegae and M. Wada (2004) DNA interference: a simple and efficient gene-silencing system for high-throughput functional analysis in the fern Adiantum. Plant Cell Physiol. 45: 1648-1657. |
|
Ichikawa, K., M. Sugita, T. Imaizumi, M. Wada and S. Aoki (2004) Differential expression on a daily basis of plastid sigma factor (PpSig) genes from the moss Physcomitrella patens: Regulatory interactions among PpSig5, the circadian clock and blue light signaling mediated by cryptochromes. Plant Physiology in press. |
Review articles |
|
Wada, M, and N. Suetsugu (2004) Plant organelle positioning. Current Opinion of Plant Biology 7:626-631. |
|
Kasahara, M. and M. Wada (2004) Chloroplast avoidance movement. In: Annual Plant Reviews “Plastids” Edited by Simon Moller, Kluwer Academic Publishers.In press. |
|
Kanegae, T. and M. Wada (2004) Photomorphogenesis of Ferns. In, Photomorphogenesis in Plants 3rd Edition Edited by Schafer and Nagy, Kluwer Academic Publishers,Dordrecht. In press. |
|
Suetsugu, N., and M. Wada (2004) Photoreceptor Gene Families in Lower Plants. In, Handbook of Photosensory Receptors. Edited by W.R. Briggs and J.L. Spudich, Wiley-VCH Verlag, Weinheim. In press. |
|
Wada, M. (2005) Chloroplast movement.In: Light Sensing in Plants, Edited by M. Wada, K. Shimazaki, and M. Iino. Springer-Verlag, Tokyo In press. |
