
 |
DIVISION FOR MORPHOGENESIS |
| Professor: | UENO, Naoto |
| Associate Professor: | KINOSHITA, Noriyuki |
| Research Associates: | NAKAMURA, Makoto |
| TAKAHASHI, Hiroki | |
| Technical Staff: | TAKAGI, Chiyo |
| Technical Assistants: | YAMAMOTO, Takamasa |
| TERASAKA, Chie | |
| MOROKUMA, Yoshie | |
| YAMADA, Shigehiro | |
| HAYASHI, Kazue | |
| SHIBAMOTO, Kaori | |
| ASANO, Megumi | |
| NIBB Research Fellow: | MIURA-HYODO, Junko |
| Postdoctoral Fellows: | KITAYAMA, Atsushi |
| MOROKUMA, Junji | |
| HOTTA, Kohji | |
| Graduate Students: | IIOKA, Hidekazu 1) |
| MIYAKOSHI, Akira 1) | |
| CHUNG, Hyeyoung 2) | |
| KUJIRAOKA, Masahiro 1) | |
| YOSHIKANE, Nami 3) | |
| MAEDA, Masato 1) | |
| GOTO, Masanori 1) | |
| LEE, Rebecca 1) | |
| Secretaries: | MIYAKE, Satoko |
| TSUGE, Toyoko | |
| 1) Graduate University for Advanced Studies | |
| 2) University of Tokyo | |
| 3) Nara Institute of Science and Technology | |
|
The complex morphogenesis of organisms is achieved by consecutive cell-to-cell interactions during development. Recent studies suggest that growth factors play crucial roles in controlling such intercellular communications in a variety of organisms. In addition to secretory factors which trigger intracellular signaling, transcription factors which act in the nucleus to regulate gene expression are thought to be essential for the determination of cell fates. Our main interest is to know how pattern formation and morphogenesis during development is regulated by these growth factors and transcription factors. We address this problem using several model animals, including frog, fly and acidian, employing embryology, genetics, molecular and cellular biology, and biochemistry. In addition, we have recently introduced genomics technologies to elucidate precise genetic program controlling early development. |
I. Gastrulation movement regulated by Wnt signaling |
|
Gastrulation is one of the most important processes during morphogenesis of early embryo, involving dynamic cell migration and change in embryo shape. Almost all animals undergo gastrulation to form the gut. In spite of its importance, the mechanism underlying the event has just begun to be studied at molecular level. During Xenopus gastrulation, mesodermal cells migrate to the inside of the embryo and move on the blastocoel roof. One of the important mechanisms for this process is the cell movement called “convergent extension”. As convergent extension begins, cells are polarized and aligned mediolaterally, followed by the mutual intercalation of the polarized cells. Recent studies suggest that several extracellualr signals are essential for the polarization, one of which is fibroblast growth factor (FGF). We therefore attempted to identify FGF target genes to understand the molecular mechanism underlying the FGF function using our Xenopus cDNA microarray. As a result, we identified a gene encoding a neurotrophin receptor homolog (NRH) and demonstrated it to be an essential component of FGF to control protrusive activity of cells undergoing gastrulation. Another pathway regulating gastrulation is one of the Wnt signaling pathways, called Wnt/JNK (c-Jun N-terminal kinase) pathway is shown to be important for the regulation of convergent extension. The pathway is highly conserved among species and initially found to be essential for the establishment of planar cell polarity (PCP) of Drosophila wing hair. We have previously demonstrated that Xenopus prickle (Xpk), a Xenopus homologue of a Drosophila PCP gene, is an essential component for gastrulation cell movement. Subsequently, we identified a protein that binds to XPK by yeast two-hybrid screening. The XPK binding protein was found to be a member of the Ste20 kinase family and named as Xenopus prickle-interacting kinase, XPIK. Loss-of-function of XPIK resulted in severe defects in gasrtulation causing spina bifida. XPIK is not only sufficient to activate JNK in embryo, but also required for full activation of JNK by Dishevelled. These suggest that XPIK also plays an essential role in connecting extracellular Wnt signal to JNK activation through Dishevelled and XPIK. In addition, we have been attempting to identify novel regulatory components controlling gastrulation cell movements by an expression cloning method based on morphology of dorsal marginal zone explant (Keller’s explant) and of embryo. After two thousand clones were examined by overexpression in the dorsal region of embryos, approximately 5% of clones were found to perturb normal gastrulation cell movements. Functional relevance of identified genes to cell movements is currently under investigation. |
II. Wnt signaling pathway regulates actin cytoskeletal dynamics during gastrulation |
|
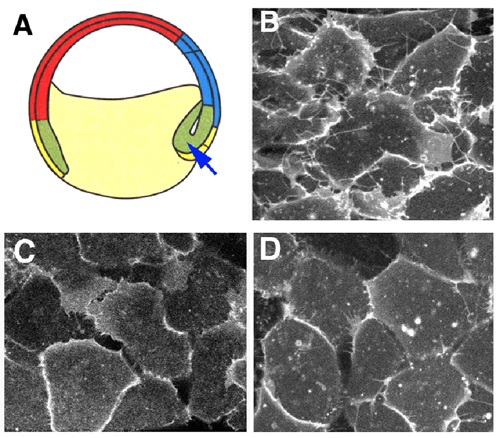
The noncanonical Wnt pathway has been implicated in the regulation of Xenopus gastrulation movements. Loss of function of its signaling components, such as Dishevelled, Rho GTPases and c-Jun N-terminal kinase, leads to a severe gastrulation defect. We investigate the molecular mechanism of how the Wnt pathway regulates this morphogenetic process. Convergent extension of the dorsal mesoderm is one of the important mechanisms during gastrulation. In this movement, cells are polarized, elongated and then, intercalated. We have established a technique to observe this process at the cellular level. This method enabled us to show that loss of function of the Wnt signaling components caused a defect in the polarization of the mesodermal cells. During convergent extension, the formation of lamelipodia- and filopodia-like protrusions from the mesodermal cells are observed. We found that the blockade of the Wnt signaling pathway significantly reduced the protrusive activity. On the contrary, overexpression of Xwnt11 and its receptor Frizzled promoted the protrusive activity. These results indicate that the noncanonical Wnt pathway plays an essential role in the convergent extension movements through the regulation of actin cytoskeletal dynamics. |
 |
|
Figure 1 The Wnt signaling pathway regulates protrusive activity in the mesodermal cells. (A)The dorsal mesodermal cells indicated by the arrow were observed. Control cells (B) form lamelipodia- and filopodia-like protrusions, whereas this activity was significantly reduced by the expression of the dominant negative mutant of Xwnt11 (C) or Dishevelled (D) |
III. One of the DPP signal downstream components TNA/TONAS participates roles in protein SUMO modification and epigenetic regulation |
|
Drosophila is one of the ideal model organisms to dissect signal transduction pathway by genetic methods. We have carried out dominant suppressor screening for a transgenic mutant fly that expresses activated DPP/BMP type-I receptors in wing imaginal discs. Two mutant alleles of the tonalli (tna) gene were isolated in this screening. tna is a previously reported mutant and a member of Trithorax group genes. Trithorax group components have been shown to be involved in the epigenetic regulation including Histone modification and chromatin remodeling. We isolated vertebrate homologues of tna and named as tonalli related SP-RING finger protein, TONAS-1 and TONAS-2. The most characteristic feature of these proteins is the existence of a single SP-RING finger motif in the middle. We have been investigating function of TNA/TONAS family SP-RING finger proteins by genetical and biochemical approarches. The SP-RING motif was originally found in the PIAS family SUMO-E3 ligase proteins. We found that TONAS can bind E2 enzyme ubc9 and has activity stimulating SUMO-2/3 specific conjugation to TONAS itself. SUMO-2/3 conjugated TONAS proteins were predominantly recovered in cytoplasmic fraction. The result indicates thata SUMO-2/3 modification somehow regulates nuclear export or cytoplasmic retention for SUMO-2/3 modified substrates. Our ongoing study also suggests that TNA/TONAS proteins participates a role in complex Histone modifications. |
IV. Function of Brachyury-downstream notochord-specific genes in morphogenetic movements of the Ciona intestinalis embryo |
|
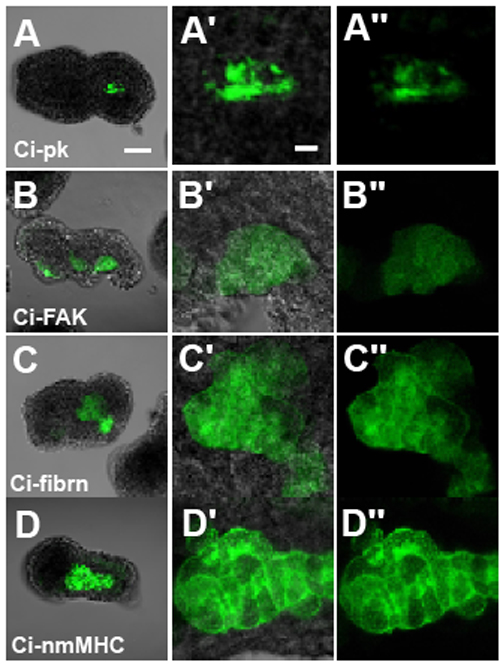
Ascidians, urochordates, are one of the three chordate groups, and the ascidian tadpole is thought to represent the most simplified and primitive chordate body plan. It contains a notochord, which is a defining characteristic of chordate embryo composed of only 40 cells. To understand the morphogenesis in this simple system, we have focused on a gene, Brachyury, which is known to play an important role in the notochord development. In ascidian, Brachyury is expressed exclusively in the notochord and the misexpression of the Brachyury gene (Ci-Bra) of Ciona intestinalis is sufficient to transform endoderm into notochord. This gene encodes a sequence-specific activator that contain a T-box DNA-binding domain, and in vertebrates, it is initially expressed throughout the presumptive mesoderm and gradually restricted to the developing notochord and tailbud. The phenotype of the Brachyury mutants in mice and zebrafish revealed that this gene is essential for notochord differentiation. Our goal is to elucidate the down stream pathway of this important gene in ascidian in order to set the stage for understanding not only the formation and function of the notochord but how this important structure has evolved. Formation of the chordate body is accomplished by a complex set of morphogenetic movements of constituent cells. Convergent extension is a major component of axis elongation. In the urochordate ascidian Ciona intestinalis, Brachyury (Ci-Bra) plays a key role in the formation of the notochord, which undergoes typical convergent extension. Nearly forty Ci-Bra downstream notochord genes have been identified. The present functional analyses of 32 genes revealed that 24 (75%) of them are involved in convergent extension; nine genes including Ci-prickle, Ci-PTP and Ci-talin caused failure of convergent extension when their function was suppressed with specific morpholinos, while the function of 15 other genes including Ci-netrin, Ci-ERM and Ci-pellino was required for intercalation/extension. In addition, knockdown of Ci-Noto7 resulted in the failure of convergence but intercalation/extension took place normally suggesting a separable phase of convergent extension. |
 |
|
Figure 2 Failure in convergent movements caused by functional suppression of four genes. (A) Ci-pk, (B) Ci-FAK, (C) Ci-fibrn, (D) Ci-nmMHC. (A-D) Whole embryos with EGFP expression, (A’-D’) enlargement of notochord region, and (A”-D”) their dark-field images. Scale bars, 50 µm in A and 10 µm in A’. |
V. Xenopus functional genomics (Xenomics) |
|
Using Xenopus laevis as a model, we have been attempting to reveal the complex regulatory network controlling the morphological processes in early embryogenesis that are associated with vigorous cell proliferation and differentiation through the clarification of gene function and their interactions in the system. In order to achieve the goal, we first collected more than 170,000 ESTs from the early embryo cDNA libraries. These ESTs were assembled into more than 30,000 contigs and singlets. Among them, we have selected full-length cDNAs excluding redundancy, and have read the full insert sequence for about 3,000 novel genes. To explore the spatiotemporal expression profile of each gene, we have carried out whole mount in situ hybridization (WISH) of embryos at different stages of early embryogenesis. So far, we have investigated the expression patterns of more than 2,000 genes. We have also started to investigate the subcellular localization of each gene product using GFP-labeled expression vector. We constructed the comprehensive database XDB3 that stores EST sequences, assembled sequences, full insert sequences and WISH data with the descriptions of presumed gene function. We also created microarray with PCR-amplified cDNAs, and carried out a series of microarray experiments on the collaboration basis. In order to investigate how the gene expression is regulated by the cell-cell interaction, we combine the microarray technology with classical embryo manipulations including surgical operations. |
 |
|
Figure 3 Xenopus Data Base 3 (http://xenopus.nibb.ac.jp). |
Publication List |
|
Sakamaki, K., Takagi, C., Kominami, K., Sakata, S., Yaoita, Y., Kubota, H. Y., Nozaki, M., Yonehara, S., and Ueno, N. (2004) The adaptor molecule FADD from Xenopus laevis demonstrates evolutionary conservation of its pro-apoptotic activity. Genes Cells. 9, 1249-1264. Chung, H. A., Hyodo-Miura, J., Kitayama, A., Terasaka, C., Nagamune, T., and Ueno, N. (2004) Screening of FGF target genes in Xenopus by microarray: temporal dissection of the signalling pathway using a chemical inhibitor. Genes Cells. 9, 749-761. Ohkawara, B., Shirakabe, K., Hyodo-Miura, J., Matsuo, R., Ueno, N., Matsumoto, K., and Shibuya, H. (2004) Role of the TAK1-NLK-STAT3 pathway in TGF-beta-mediated mesoderm induction. Genes Dev. 18, 381-386. Iioka, H., Ueno, N., and Kinoshita, N. (2004) Essential role of MARCKS in cortical actin dynamics during gastrulation movements. J Cell Biol. 164, 169-74. Miyakoshi, A., Ueno, N., and Kinoshita, N. (2004) Rho guanine nucleotide exchange factor xNET1 implicated in gastrulation movements during Xenopus development. Differentiation. 72, 48-55. Kawai, N., Takahashi, H., Nishida, H., and Yokosawa, H. (2005) Regulation of NF-kappaB/Rel by IkappaB is essential for ascidian notochord formation. Dev Biol. 277, 80-91. Takatori, N., Hotta, K., Mochizuki, Y., Satoh, G., Mitani, Y., Satoh, N., Satou, Y., and Takahashi, H. (2004) T-box genes in the ascidian Ciona intestinalis: characterization of cDNAs and spatial expression. Dev Dyn. 230, 743-753. Uchimura, Y., Nakamura, M., Sugasawa, K., Nakao, M., and Saitoh, H. (2004) Overproduction of eukaryotic SUMO-1- and SUMO-2-conjugated proteins in Escherichia coli. Anal Biochem. 331, 204-206. |
