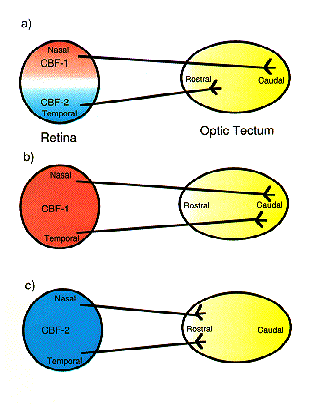
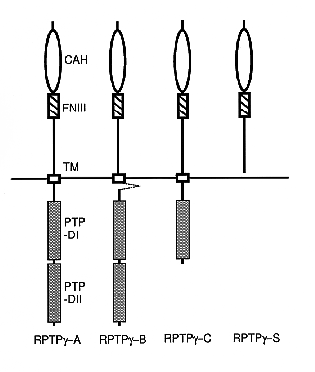
NATIONAL INSITUTE FOR BASIC BIOLOGY

National Institute for Basic Biology
Division of Molecular Neurobiology
- Professor:
- Masaharu Noda
- Research Associates:
- Nobuaki Maeda
- Masahito Yamagata
- Eiji Watanabe
- Masahito Yamagata
- NIBB Fellow:
- Masakazu Takahashi
- Graduate Students:
- Hiroki Hamanaka (- Sept. 30, 1996)
- Junichi Yuasa
- Takafumi Shintani
- Taeko Nishiwaki
- Chika Saegusa (Oct. 1, 1996 -)
- Junichi Yuasa
- Visiting Scientists:
- Ikuko Watakabe (Oct. 1, 1996 -)
- Tatsunori Yamamoto* (- Aug. 31, 1996)
- Technical Staffs:
- Akiko Kawai
- Shigemi Ohsugi