10.0Ѓ@ Retrieving Genomic Sequences for Gene
Targeting Using the Available Web-Based Browsers
Kari Thompson and Yuji Hiwatashi
Introduction
In
order to construct expression and knockout moss lines first you must obtain DNA
fragments located in the 5' and 3' regions of a targeting coding sequence. The
size of the DNA fragments is usually between 1 and 2 kb.Ѓ@
There
are two websites; you can use to get these sequences:
JGI:
http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html
Physcobase:
http://moss.nibb.ac.jp/
1.)
Using JGI to retrieve genomic sequence:
Procedure:
1.) Click on the BLAST tab.
2.) Choose the alignment
program blastn: blast nucleotide vs. nucleotide.
3.) Leave the defaults for
expect and word size OK for beginning.
4.) Choose the database Physcomitrella
patens v1.1 repeat masked main genome sequence.Ѓ@
5.) Paste the genomic or cDNA
sequence of your gene of interest into the box for query sequence.Ѓ@
6.) You may enter your e-mail
address if you would like the result e-mailed to you, but it is not necessary.
7.) Click Submit job.
8.) Please wait while the
server searches for sequences that show similarity.Ѓ@ This may take a few minutes depending on queries ahead of yours.
9.) The output will show you
scaffolds that have similarity to your sequence in a graphical format.Ѓ@ Scaffolds that are red show the most
similarity to your sequence, the first scaffold likely contains your gene.Ѓ@ Make a note of the scaffold number.
10.) Mouse over the graph and
click on the red line for the scaffold.Ѓ@
11.) At the top of the screen
you will see a table.Ѓ@ This contains
your query length, the scaffold number, and where your query sequence lies in
the scaffold.Ѓ@
For example:
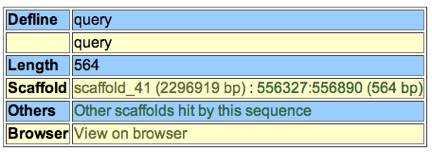
This means that your query length
was 564 bp and it is located in scaffold 41 between 556327 and 556890 bp.Ѓ@ It is best to make a note of the location of
your cDNA sequence; it will be useful in the following steps.Ѓ@ You can click on others to view other
scaffolds that contain your gene.Ѓ@
Beneath the table you can see a
graph that shows the similarity between your sequence and the scaffold. Ѓ@
For example:
Ѓ@Ѓ@Ѓ@Ѓ@Ѓ@ Below this you can see the alignment of your query and the
scaffold, if you click on seq next to scaffold seq you can retrieve the
scaffold sequence the corresponds to your query.Ѓ@
12.) Click on the scaffold
name found in the first table (shown in step 11).Ѓ@ In this example you would click on scaffold_41.
13.) In this table you can
see all of the contigs that were used to assemble the scaffold.Ѓ@
14.) You can click on get
sequence
to retrieve the entire genomic sequence for the scaffold, but it is easier to
scroll down and find the contig that contains the sequence of your cDNA.Ѓ@ In this case find the contig that contains
the fragment mentioned in the first table: from 556327 to 556890 bp (shown in
step 11).Ѓ@ This contig should contain
enough genomic information for you to design your desired constructs.Ѓ@ If your gene is located at the beginning or
the end of the contig you may need to get the entire sequence to make a good
judgment of the genomic sequence.Ѓ@
2.)
Using PHYSCObase
to retrieve genomic sequence.Ѓ@
ЃgPHYSCObaseЃh http://moss.nibb.ac.jp/
ЃgBlast Assemble Data SubmissionЃh
http://moss.nibb.ac.jp/cgi-bin/blast-assemble
If
you cannot find a genomic DNA sequence corresponding to your gene by blast
searching with the JGI database, you should try to identify the sequence using ЃgBlast
Assemble Data SubmissionЃh
Procedure:
1.) Click on the link, or use
this address: http://moss.nibb.ac.jp/cgi-bin/blast-assemble.Ѓ@ Or if you start
at the homepage for PHSYCObase, click ЃgDNA databaseЃh
and then click ЃgBLAST raw WGS sequence and assemble into contigЃh
2.) You
may enter the name of your gene in the box for sequence name if you wish, but
it is not necessary.
3.) Paste the genomic
or cDNA
sequence (fasta format) of your gene of interest into the
box for sequence; this is your query.Ѓ@
4.) Choose
ЃgnucleotideЃh and ЃgPhyscomitrella patensЃh.
5.) Click Construct
Contigs.
6.) Please wait while the
server searches for sequences that show similarity.Ѓ@ This may take about 10 minutes depending on queries ahead of yours.
7.) After
a few minutes, click get the highest score scaffold
to retrieve a genomic DNA sequence corresponding to your gene.
8.) Usually
you can see three sequences in the result. The first and third sequences are
represented by lowercase letters above or under ЃennnnЃcnnnnЃf,
respectively.Ѓ@ These represent possible,
but not guaranteed, extreme 5Ѓf and 3Ѓf genomic sequences of your gene. Ѓ@The second sequence is shown in capital
letters betweenЃ@ ЃennnnЃcnnnnЃf.
The second sequence should be overlapping with the sequence that you used as
query and is usually enough for designing primers to make constructs.
9.) Copy
and paste the second sequence into a new file and use it to construct a contig
with your original query sequence.
Ѓ@Ѓ@Ѓ@Ѓ@Ѓ@