
 |
DIVISION OF EVOLUTIONARY IOLOGY |
| Professor: | HASEBE, Mitsuyasu |
| Associate Professor: | MURATA, Takashi |
| Research Associates: | FUJITA, Tomomichi |
| HIWATASHI, Yuji | |
| Technical Staff: | SUMIKAWA, Naomi |
| NIBB Research Fellow: | NISHIYAMA, Tomoaki |
| JSPS Research Fellow: | SATO, Yoshikatsu |
| Postdoctoral Fellows: | AONO, Naoki |
| MIYAZAKI, Saori | |
| Graduate Students: | HASHIMOTO, Kaoru |
| MORINAGA, Shin-ichi | |
| (Tohoku Univ.) (Apr. 1 -) | |
| HOSOKAWA, Kentaro | |
| (Tokyo Univ.) (Apr. 1 -) | |
| Technical Assistants: | AOKI, Etsuko |
| BITOH, Yoshimi (- Apr. 30) | |
| NARUSE, Mayumi (- Jun. 30) | |
| HAYAKAWA, Yuko (Aug. 1-31) | |
| ICHIKAWA, Yuki (Jul. 16 -) | |
| KAJIKAWA, Ikumi (Aug .1-31) | |
| KITANI, Masakazu | |
| MAKINO, Haruko (May. 1 -) | |
| OONO, Chikako (- Jun. 30) | |
| SUZUKI, Yoriko | |
| TANIKAWA, Yukiko (- Jun. 30) | |
| WATASE, Masataka (May. 1 -) | |
| Secretaries: | KABEYA, Kazuko (- Mar. 30) |
| KOJIMA, Yoko (May. 1 -) |
|
All living organisms evolved from a common ancestor that lived more than 3.5 billion years ago, and the accumulation of mutations in their genomes has resulted in the present biodiversity. Traces of the evolutionary process are found in the genomes of extant organisms. By comparing the gene sequences and gene networks of different organisms, we can infer (1) the phylogenetic relationships of extant organisms and (2) the genetic changes that caused the evolution of morphology and development. The inferred phylogenetic relationships provide important insights into problems in various fields of evolutionary biology. Our group focuses on biogeography, the evolution of morphological traits, and systematics in a wide range of taxa. Concerning the evolution of morphology and development, we hope to explore the genetic changes that led to the evolution of the plant body plan. We have selected Arabidopsis (angiosperm), Gnetum (gymnosperm), Ginkgo (gymnosperm), Ceratopteris (pteridophyte), Physcomitrella (bryophyte), and some green algae as models to compare the functions of genes involved in the development of both reproductive and vegetative organs in land plants. |
I. Origin of the Plant CellThe first green alga cell evolved via symbiosis between an ancestral non-photosynthetic eukaryote and a cyanobacterium. Cyanobacteria now exist as chloroplasts in the host cell. The factors and mechanisms of chloroplast movement are being investigated to reveal the molecular mechanisms used to"domesticate" cyanobacteria as organelles. Analyses of (1) cytosolic calcium icon concentration and cytoskeleton organization during chloroplast movement in the moss Physcomitrella patens and (2) the functional divergence of photoreceptors and motor proteins involved in chloroplast movement in the moss from angiosperms are in progress by a team directed by Y. Sato. |
II. Evolution from unicellular to multicellular organismsThe first evolutionary step from unicellular to multicellular organisms is to form two different cells from a single cell via asymmetric cell division. The first cell division of a protoplast isolated from the protonemata of the moss Physcomitrella patens is asymmetric regarding to its shape and nature, and gives rise to an apical meristematic cell and a differentiated non-meristematic cell. A systematic overexpression screening for genes involved in asymmetric cell division of protoplasts in P. patens is in progress by a team directed by T. Fujita. After eliminating genes that are not directly involved in asymmetric cell divisions, such as photosynthesis,genes, we used 3000 clones as materials for the overexpression screening. Individual cDNAs were subcloned under a constitutive promoter and introduced into the protoplasts of P. patens for transient expression. We observed and categorized phenotypes of the regenerating protoplasts. Thus far we identified 58 cDNAs, whose overexpression caused the defects in asymmetric cell divisions in two repeated experiments. Overexpression of the genes in protoplasts with GFP-tubulin or GFP-talin, expression analyses of each gene-cytrin fusion protein under its native promoter, loss of function experiments using RNAi are now in progress to characterize what processes these genes are involved in. Functional analyses of these genes should help us to understand molecular mechanisms of how plants generate distinct cell lineages to build their multicellular bodies. |
III. Evolution from cells to tissuesThe most prominent difference between plant and animal cells is that plant cells have a cell wall and do not move during development. Therefore, the plane of cell division and the direction of cell elongation, which are regulated by cortical microtubules, determine the morphology of differentiated tissues and organs. |
Organization of microtubulesCortical arrays of microtubules are essential for morphogenesis in plants. We found that, by live imaging, microtubules in the arrays are formed as branches on pre-existing microtubules. γ-tubulin, a protein that is essential for the formation of microtubules in animal cells, is located at the branching point and in the cytoplasm, and a loss of γ-tubulin due to gene silencing causes a malformed organ with irregularly shaped cells. In vitro experiments using isolated plasma membrane/microtubule complexes suggested that γ-tubulin. Once cortical microtubules are formed, they can turnover without microtubules from the nucleus. Factor(s) responsible for attachment of γ-tubulin onto the side of microtubules is a key element responsible for the difference between plant and animal cells. Isolation of the factor(s) responsible for attachment of γ-tubulin onto the side of microtubules by biochemical and other approaches is in progress by a team directed by T. Murata. |
IV. Evolution of molecular mechanisms in the development of vegetative organs |
Meristem initiation and maintenancePostembryonic growth of land plants occurs from the meristem, a localized region that gives rise to all adult structures. Meristems control the continuous development of plant organs by balancing the maintenance and proliferation of stem cells, and directing their differentiation. Meristem initiation and maintenance is a fundamental question in plant development research. Three lines, exhibiting reporter gene (uidA) expression preferentially in the apical cells, were isolated from previously established gene- and enhancer-trap lines, and identified as encoding kinesin- and ubiquitin-like proteins, and an unknown protein. Functional analyses of these genes are currently under investigation by a team directed by Y. Hiwatashi. The disruption of the kinesin-like gene did not show any phenotypic differences from the wild type. This is likely caused by the functional redundancy of closely related genes, and the analyses of double disruptions are in progress. Disruption of the gene encoding ubiquitin-like protein suggests that the gene is involved in cell division and elongation through microtubule organization with the proteasome complex. |
Evolution of shoot meristemThe angiosperm shoot apical meristem is a dome of small, proliferating cells whose organization is highly structured into layers and/or zones. The apical meristem of many seed-less vascular plants contains a single apical cell or initial that is apparent in both the gametophyte (haploid) and sporophyte (diploid) generations. Given the importance of the apical meristem in elaborating the three dimensional body plan of plants, the apical meristem is thought to be among the most important innovations for the evolution of land plants. In order to gain insights into the molecular mechanisms underlying the development and evolution of the plant meristem, we have identified and analyzed the expression of two class 1 and one class 2 KNOX (knotted-like homeobox) genes from the fern Ceratopteris richardii. Expression of both class 1 genes was detected in the shoot apical cell, leaf primordia, marginal part of the leaves, and vascular bundles by in situ hybridization, a pattern that closely resembles that of class 1 KNOX genes in angiosperms with compound leaves. The fern class 2 gene was expressed in all sporophytic tissues examined, which is characteristic of class 2 gene expression in angiosperms. Unexpectedly, all three CRKNOX genes were not detected in gametophyte tissues by RNA gel blot analysis. Arabidopsis plants overexpressing the fern class 1 genes resembled plants that overexpress seed plant class 1 KNOX genes in leaf morphology. Ectopic expression of the class 2 gene in Arabidopsis did not result in any unusual phenotypes.Taken together with phylogenetic analysis, our results suggest that 1) the class 1 and 2 KNOX genes diverged prior to the divergence of fern and seed plant lineages; 2) the class 1 KNOX genes function similarly in seed plant and fern sporophyte meristem development despite their differences in structure, 3) KNOX gene expression is not required for the development of the ferngametophyte, and 4) the sporophyte and gametophytic meristems of ferns are not regulated by the same developmental mechanisms at the molecular level (Sano et al. in press). To further investigate the evolution of meristem and KNOX genes, characterization of the moss Physcomitrella KNOX genes are in progress by a team directed by T. Nishiyama. |
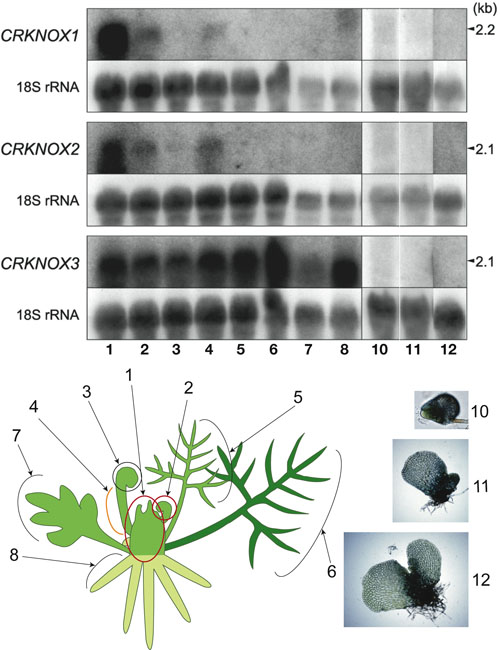
|
|
(Figure 1) Expression patterns of three KNOX genes in the fern Ceratopteris richardii. The tissues from which RNAs were extracted are shown in the schematic figures. |
V. Origin and evolution of floral homeotic genesThe flower is the reproductive organ in angiosperms, and floral homeotic genes, such as MADS-box genes and FLO/LEAFY genes, regulate floral organ identity. To investigate the origin of floral homeotic genes, the functions of these genes in several basal angiosperms, the moss Physcomitrella, and three green algae (Chara, Coleochaete, and Closterium) are being analyzed. Land plants are believed to have evolved from a gametophyte-dominant ancestor without a multicellular sporophyte; most genes expressed in the sporophyte probably originated from those used in the gametophyte during the evolution of land plants. To analyze the evolution and diversification of MADS-box genes in land plants, eight MADS-box genes predominantly expressed in pollen, male gametophyte, are analyzed by a team directed by N. Aono. |
VI. Molecular mechanisms of speciationReproductive isolation is the first step in speciation. To obtain insights into reproductive isolation, several receptors specifically expressed in the pollen tube are being studied to screen for the receptors involved in pollen tube guidance by a team directed by S. Miyazaki. Polyploidization is a major mode of speciation in plants, although the changes that occur after genome duplication are not well known. Polyploid species are usually larger than diploids, but the mechanisms responsible for the size difference are unknown. To investigate these mechanisms, tetraploid Arabidopsis was established and its gene expression patterns are being compared to those of diploid wild-type plants using microarrays. |
VII. Phylogenetic analysis of land plantsOpinions on the basal relationship of land plants vary considerably and no phylogenetic tree with significant statistical support has been obtained. We performed phylogenetic analyses using 51 genes from the entire chloroplast genome sequences of 20 representative green plant species. The analyses, using translated amino acid sequences, indicated that extant bryophytes (mosses, liverworts, and hornworts) form a monophyletic group with high statistical confidence, and that extant bryophytes are likely sister to extant vascular plants, although the support for monophyletic vascular plants was not strong. Analyses at the nucleotide level could not resolve the basal relationship with statistical confidence. Bryophyte monophyly inferred using amino acid |
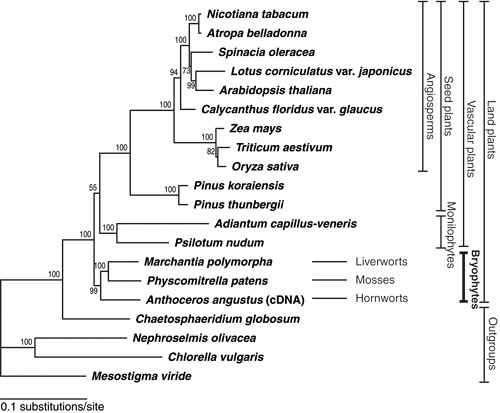
|
|
(Figure 2) Phylogenetic relationship of representative lineages in land plants inferred using 8,979 amino acid residues for 51 genes from 20 chloroplast genomes. sequences has a good statistical foundation and is not rejected statistically by other datasets. We proposed bryophyte monophyly as the current best hypothesis (Nishiyama et al. 2004). |
VIII. Evolution of RNA editing in land plantsWe sequenced transcripts from all putative genes for proteins, rRNAs, and a selection of those encoding tRNAs in the chloroplast genome of the fern Adiantum capillus-veneris. We detected 349 RNA editing sites when the cDNA sequence was compared to that of the genomic DNA. The level of RNA editing in this fern is more than ten times that of any other vascular plant examined across an entire chloroplast genome. A previous study found even higher levels of editing in a hornwort (942 sites). This suggests that the relatively low levels of editing in seed plants (less than 0.05%) may not be typical for land plants, and that RNA editing may play a major role in chloroplast genome processing. Additionally, we found that 53 editing sites in Adiantum are homologous to editing sites in the hornwort, and some other land plants. This implies that a major component of RNA editing sites have been conserved for hundreds of millions of years. |
Publication list: |
|
Sano, R., Juárez, C. M., Hass, B., Sakakibara, K., Ito, M., Banks, JA, and Hasebe, M. 2005. KNOX class of homeobox genes potentially have similar function in both diploid unicellular and multicellular meristems, but not in haploid meristems. Evol. Dev. in press. |
|
Kishi, M., Murata, T., Hasebe, M., and Watanabe, Y. 2005. An extraction method for tobacco mosaic virus movement protein localizing in plasmodesmata. Protoplasma in press. |
|
Hattori, M., Hasebe, M., and Sugita, M. 2004. Identification and characterization of cDNAs encoding pentatricopeptide repeat (PPR) proteins in the earliest land plant, the moss Physcomitrella patens. Gene 343: 305-311 |
|
Tamura, M.N., Fuse, S., Azuma, H., and Hasebe, M. 2004. Biosystematic studies on the family Tofieldiaceae I. Phylogeny and circumscription of the family inferred from DNA sequences of matK and rbcL. Plant Biol. 6: 562-567. |
|
Rutherford, G., Tanurdzic, M., Hasebe, M., and Banks, J.A. 2004. A systemic gene silencing method suitable for high throughput, reverse genetic analyses of gene function in fern gametophytes. BMC Plant Biology 4: 6. |
|
Wolf, P.G., Rowe, C.A., and Hasebe, M. 2004. High levels of RNA editing in a vascular plant chloroplast genome: analysis of transcripts from the fern Adiantum capillus-veneris. Gene 339: 89-97. |
|
Nishiyama, T., Wolf, P.G., Kugita, M., Sinclair, R.B., Sugita, M., Sugiura, C., Wakasugi, T., Yamada, K., Yoshinaga, K. Yamaguchi, K., Ueda, K., and Hasebe, M. 2004. Chloroplast,phylogeny indicates that bryophytes are monophyletic. Mol. Biol. Evol. 21: 1813-1819. |
|
Aoki, S., Uehara, K., Imafuku, M., Hasebe, M., and Ito, M. 2004. Phylogeny and divergence of basal angiosperms inferred from APETALA3- and PISTILLATA-like MADS-box genes. J. Plant Res. 117: 229-244. |
