| RESEARCH SUPPORT FACILITY | ||
|
||
| |
||
| I. Facilities 1. The Large Spectrograph Laboratory 1. The Large Spectrograph Laboratory This laboratory provides, for cooperative use, the Okazaki Large Spectrograph (OLS), which is the largest spectrograph in the world, dedicated to action spectro-scopical studies of various light-controlled biological processes. The spectrograph runs on a 30kW Xenon arc lamp and has a compound grating composed of 36 smaller individual gratings. It projects a spectrum of a wavelength range from 250nm (ultraviolet) to 1,000nm (infrared) onto its focal curve of 10m in length. The fluence rate (intensity) of the monochromatic light at each wavelength is more than twice as much as that of the corresponding monochromatic component of tropical sunlight at noon (Watanabe et al. 1982, Photochem. Photobiol., 36, 491-498). An advanced irradiation system composed of CW lasers (364nm, 390-410nm,
440-460nm, 532nm, 655nm, 752nm) and uniform-fluence-rate irradiation
optics interconnected by optical fibers was constructed in 2003. An
advanced observation system for cellular and intracellular photobiological
responses utilizing a two-photon microscope (FV300-Ix71-TP with a
MaiTai laser) and a microbial photomovement analyzer (WinTrack2000/Ecotox)
etc. was also introduced. Various equipments for tissue and cell culture are provided. This
laboratory is equipped with safely rooms which satisfy the P2/P3 physical
containment level. This facility is routinely used for DNA recombination
experiments. Computer laboratory maintains several computers to provide computation resources and means of electronic communication in this Institute. Currently, the main system consists of three servers and two terminal work-stations: biological information analysis server (SGI Ori-gin 2000), database server (Sun Enterprise 450), file server (Sun Enterprise 3000), data visualization terminal and molecular simulation terminal (both are SGI Octanes). Some personal computers and color/monochrome printers are also equipped. On this system, we provide various biological databases and data retrieval/analysis programs, and support large-scale data analysis and database con-struction for the Institute members. Computer laboratory also provides network communi-cation services
in the Institute. Most of PCs in each laboratory as well as all of
the above service machines are connected each other with local area
network (LAN), which is linked to the high performance multimedia
back-bone network of Okazaki National Research Institute (ORION).
Many local services including sequence analysis service, file sharing
service and printer service are provided through this LAN. We also
maintain a public World Wide Web server that contains the NIBB home
pages (http://www.nibb.ac.jp). There are a large number of culture boxes, and a lim-ited number of
rooms with environmental control for plant culture. In some of these
facilities and rooms, ex-periments can be carried out at the P1 physical
contain-ment level under extraordinary environments such as strong
light intensity, low or high temperatures. This laboratory consists of two 20 m2 glass-houses with
precise temperature and humidity control, three green houses (each
6 m2) at the P1 physical containment level, a small farm,
two greenhouses (45 and 88 m2) with automatic sprinklers.
The laboratory also includes a building with storage and work space. Autotrophic and heterotrophic culture devices and equipment for experimental
cultures of plant and micro-bial cells in this laboratory. A facility
for preparation of plant cell cultures including an aseptic room with
clean benches, is also provided. This laboratory was found to study transgenic plants with respect
to tolerance toward various environmental stresses. It is located
in the Agricultural Experimental Station of Nagoya University (30
km from National In-stitute for Basic Biology). The laboratory provides
a vari-ety of growth chambers that precisely control the condi-tions
of plant growth and dacilities for molecular biologi-cal and physiological
enaluations of transgenic plants. The faculty of the Research Support Facility conducts its own research
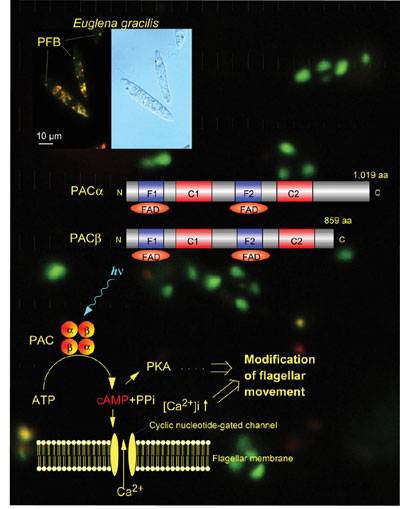
as well as scientific and administrative public services. A novel blue-light receptor with an effector role was found from Euglena gracilis (Fig. ; Iseki et al. 2002, Nature 415, 1047-1051): Euglena gracilis, a unicellular flagellate, shows blue-light type photomovements. The action spectra indicate the involvement of flavoproteins as the photoreceptors mediating them. The paraflagellar body (PFB), a swelling near the base of the flagellum has been considered as a photosensing organelle for the photomovements. To identify the photoreceptors in the PFB, we isolated PFBs and purified the flavoproteins therein. The purified flavoprotein (ca. 400 kDa), with noncovalently bound FAD, seemed to be a heterotetramer of a- and b-subunits. Predicted amino acid sequences of each of the subunits were similar to each other and contained two FAD-binding domains each followed by an adenylyl cyclase catalytic domain. The flavoprotein showed an adenylyl cyclase activity, which was elevated by blue-light irradiation. Thus, the flavoprotein (PAC, photoactivated adenylyl cyclase) can directly transduce a light signal into a change in the intracellular cyclic AMP level without any other signal transduction proteins. The involvement of PAC in positive and negative phototaxis (steering
response with respect to stimulus light direction) was also demonstrated
by its knock-down using RNAi (Ntefidou et al. 2003). Orthologues
of PAC a and b were detected in several relatives of Euglena and their phylogenetic
analysis indicated that they were trtansfered to euglenoids on the
occasion of secondary endosymbiosis (Koumura et al. submitted).
In addition to these developments, we are also trying to apply the
computational methods to real data analyses, especially to the comparative
genomics with some closely related organisms. In collabolation with
Dr. Takami's group (JAMSTEC), we are comparing genomic sequences of
some Bacillus-related organisms that live in several different environmental
conditions. The NIBB Cooperative Research Program for the Use of the OLS supports
about 20 projects every year conducted by visiting scientists including
foreign scien-tists as well as those in the Institute. |