| DIVISION OF GENE EXPRESSION AND REGULATION I | ||||||
|
||||||
* from October, 2003 ** until March, 2003 *** from June, 2003 **** from April until November, 2003 ***** from November, 2002 1) Graduate University for Advanced Studies 2) from Graduate School of Tokyo University 3) from Graduate School of Hokkaido University |
||||||
The main interest of the group is in
understanding the biology of the dynamic genome, namely, genome organization
and reorganization and its impact on gene expression and regulation.
We are also characterizing various aspects of genetic and epigenetic
gene regulations particularly on flower pigmentation of morning glories.
In addition, we are undertaking reverse genetic approaches in order
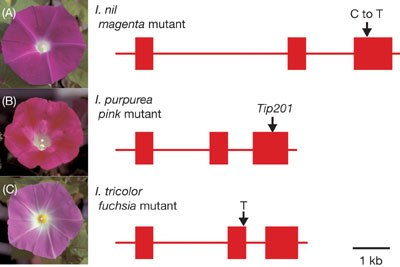
to elucidate the nature of dynamic genome in plants. Considerable attention has recently been paid to the morning glory genus Ipomoea because of the experimental versatility of its floral biology including the genetics of floral variation, flavonoid biosynthesis, and transposon-induced mutations. The genus Ipomoea includes about 600 species distributed on a worldwide scale that exhibit various flower morphologies and pigmentation patterns. A large number of Ipomoea species can be found in the Americas, particularly in Mexico. Among the genus Ipomoea, three morning glories, Ipomoea nil (the Japanese morning glory), Ipomoea purpurea (the common morning glory), and Ipomoea tricolor, were domesticated well as floricultural plants, and many mutants displaying various flower pigmentation patterns were isolated. Among these morning glories, I. nil and I. purpurea belong to the same subgenera Ipomoea, whereas I. tricolor was classified into another subgenera, Quamoclit. Both I. nil and I. tricolor display blue flowers (see Figure 2A) that contain the peonidin (3’ methoxyl cyanidin) derivative named Heavenly Blue Anthocyanin (HBA), and I. purpurea produces dark purple flowers containing a cyanidin derivative that lacks one glucose molecule and a methyl residue from HBA. All of them produce dark-brown seeds. As floricultural plants, spontaneous mutants of I. purpurea and I. tricolor displaying either white or reddish flowers were obtained while various spontaneous mutants of I. nil exhibiting many different flower colorations were generated. Although I. nil has long been believed to have originated from Southeast Asia, a hypothesis that I. nil may have arrived in Asia from tropical America was also recently proposed. In either case, the plant had been introduced into Japan from China approximately in the 8th Century as a medicinal herb. The seeds were used as a laxative, and the plant became a traditional floricultural plant in Japan around the 17th Century. I. nil has an extensive history of genetic studies, and a number of its spontaneous mutants related to the color and shape of the flowers have been isolated. It was also used extensively for physiological studies of the photoperiodic induction of flowering. The wild-type I. purpurea plant, which originated from Central America, was probably introduced to Europe in the 17th Century, and cultivars with white and red flowers were already recorded in the late 18th Century. Like I. purpurea, I. tricolor originated from Central America, and several spontaneous mutants exhibiting various flower pigmentations were obtained in the mid-20th Century. As Figure 1 shows, the spontaneous mutants of I. nil, I. purpurea, and I. tricolor carrying the magenta, pink, and fuchsia alleles, respectively, produce reddish flowers containing pelargonidin derivatives, and all of them are deficient in the gene for flavonoid 3’-hydroxylase (F3’H). The magenta allele in I. nil is a nonsense mutation caused by a single C to T base transition generating the stop codon TGA, and the cultivar Violet carries the same mutation. Several tested pink mutants in I. purpurea carry inserts of the 0.55-kb DNA transposable element Tip201 belonging to the Ac/Ds superfamily at the identical site. No excision of Tip201 from the F3’H gene could be detected, and both splicing and polyadenylation patterns of the F3’H transcripts were affected by the Tip201 integration. The fuchsia allele in I. tricolor is a single T insertion generating the stop codon TAG, and the accumulation of the F3’H transcripts was drastically reduced by the nonsense-mediated RNA decay. The I. tricolor spontaneous mutant Blue Star carrying the
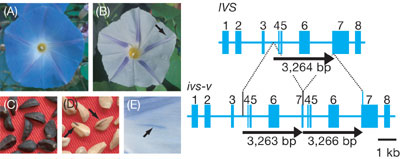
mutable ivory seed-variegated allele exhibits pale-blue flowers
with a few fine blue spots and ivory seeds with tiny dark-brown spots
(Figure 2). The mutable allele is caused by an intragenic tandem duplication
of 3.3 kb within a gene for transcriptional activator containing a
bHLH DNA-binding motif. Each of the tandem repeats is flanked by a
3-bp sequence AAT, indicating that the 3-bp microhomology is used
to generate the tandem duplication. The transcripts in the pale-blue
flower buds of the mutant contain an internal 583-bp tandem duplication
that results in the production of a truncated polypeptide lacking
the bHLH domain. The mRNA accumulation of most of the structural genes
encoding enzymes for anthocyanin biosynthesis in the flower buds of
the mutant was significantly reduced. The transcripts identical to
the wild-type mRNAs for the transcriptional activator were present
abundantly in blue spots of the variegated flowers, whereas the transcripts
containing the 583-bp tandem duplication were predominant in the pale-blue
background of the same flowers. The flower and seed variegations are
caused by somatic homologous recombination between an intragenic tandem
duplication in the bHLH gene, whereas various flower variegations
in Ipomoea are known to be caused by excision of DNA transposons inserted
into pigmentation genes.
Rice (Oryza sativa L.) is an important staple food for more
than half of the world’s population and a model plant for other
cereal species. We have developed a large-scale Agrobacterium-mediated
transformation procedure with a strong positive-negative selection
and succeeded in efficient and reproducible targeting of the Waxy
gene by homologous recombination without concomitant occurrence of
ectopic events, which must be an important first step for developing
a precise modification system of the genomic sequences in rice. By
improving our transformation procedure further, we are attempting
to modify Adh genes, which belong to a small multigene family
and reside adjacent to repetitive retroelements. Leaves of seedlings in the virescent mutant of rice are initially
pale yellow green due to partial deficient in chlorophyll and gradually
become green with the growth of the mutant. We have been characterizing
a spontaneous mutable virescent allele pale yellow leaf-variegated
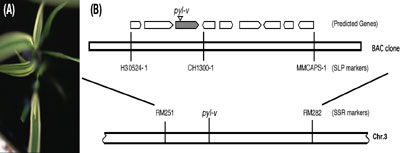
(pyl-v), conferring pale yellow leaves with dark green sectors
in its seedlings (Figure 3). The pyl-v mutant was isolated
among progeny of a hybrid between indica and japonica
rice plants. The leaf variegation is regarded as a recurrent somatic
mutation from the recessive pale yellow allele to the dark green revertant
allele. The availability of the genomic sequences of both japonica
and indica subspecies facilitates map-based cloning of the
pyl-v allele. Mapping data indicate the mutation resides
in the short arm of the chromosome 3, and excision of a new DNA transposon
from the pyl gene appears to be responsible for conferring
the leaf variegation. It is important to emphasize here that tissue
culture is necessary in all of the currently available rice reverse
genetic approaches. No somaclonal variation is likely to occur in
mutant lines induced by our newly characterizing endogenous element,
because no tissue culture has been involved in its activation. Using
a newly identified DNA transposon, therefore, we are attempting to
develop a novel transposon tagging system in rice.
|
||||||
| Publication List: Kikuchi, T., Nishimura, M., Hoshino, A., Morita, Y., Iida, S., Saito, N. and Honda, T. (2003) An efficient conversion of catechine into 3,4-trans-leucocyanidin. Heterocycles 60, 1469-1475. Hoshino, A., Morita, Y., Choi, J.D., Saito, N., Toki, K., Tanaka,Y. and Iida, S. (2003) Spontaneous mutations of the flavonoid 3’-hydroxylase gene conferring reddish flowers in the three morning glories. Plant Cell Physiol. 44, 990-1001. Yoshida, H., Akimoto, H., Yamaguchi, M., Shibata, M., Habu, Y., Iida, S. and Ozeki, Y. (2004) Alteration of methylation profiles in distinct cell lineages of the layers during vegetative propagation in carnation (Dianthus caryophyllus). Euphytica (in press). Terada, R., Asao, H. and Iida, S. (2004) A Large-scale Agrobacterium-mediated transformation procedure with a strong positive-negative selection for gene targeting in rice (Oryza sativa L.). Plant Cell Reports (in press). Iida, S., Morita, Y., Choi, J.D., Park, K.I. and Hoshino, A. (2004) Genetics and epigenetics in flower pigmentation associated with transposable elements in morning glories. Advances in Biophysics (in press). |