| DIVISION OF BIOENERGETICS | ||||||||||
|
||||||||||
1) Study abroad from Mar. 2003 2) Graduate University for Advanced Studies 3) KOBE Univ. |
||||||||||
This division aims to understand the autophagy in
respects to its molecular mechanism and its physiological role in
yeast and higher eukaryotes. Cells execute degradation processes of
their constituents together with biosynthetic processes. These two
processes are well coordinated to regulate the biological activities.
In other word, we must shed light on degradation process to fully
understand the cell, because the study on the degradation has been
retarded compared to the biosynthetic process. Autophagy is a major
route for bulk degradation of cytoplasmic constituents and organelles
in lysosome/vacuole, and is well conserved in eukaryotes. Upon nutrient starvation, autophagic process starts as building up
a membrane structure, an autophagosome, in the cytoplasm. The autophagosome
sequesters cytosol and organelles nonselectively. Then it is delivered
to the vacuole/lysosome, and the cytoplasmic materials inside are
degraded by vacuolar/lysosomal hydrolases. We had discovered autophagy
in a simple model organism, S. cerevisiae and morphologically and
genetically defined the whole process. In yeast, S. cerevisiae, Atg8 plays an important role during
autophagosome formation. We have previously reported that the Atg8
is covalently attached to phosphatidylethanolamine (PE) via a ubiquitin-like
conjugation system. The C-terminal Arg of newly synthesized Atg8 (Atg8R117)
is removed by Atg4 protease to expose a Gly residue at the C-terminus
(Atg8G116). The Apg8G116 is then activated by
Atg7 (E1 enzyme) and transferred to Atg3 (E2 enzyme). Following these
reactions, the Apg8G116 conjugates to PE through an amide
bond between its C-terminal Gly and the amino group of PE. The subsequent
deconjugation reaction by Atg4 is necessary for the normal progression
of autophagy. We developed in vitro Atg8-PE reconstitution
system. The Atg8-PE was successfully reconstituted simply with Atg8G116,
Atg7 and Atg3 by using in vivo in E. coli and in
vitro system. These results suggested that Atg7 and Atg3 are
necessary and sufficient for the Atg8-PE conjugation reaction. The
in vitro Atg8-PE reconstitution system using recombinants
and liposomes demonstrated that the efficiency of Atg8-PE conjugation
was strongly affected by lipid composition. Further, the Atg8 was
linked to the PE in liposomes, but not to the PE in the presence of
detergent, suggesting that the lipid bilayer of membrane is essential
for the Atg8-PE conjugation. Atg12 is activated by Atg7, transferred to Atg10 and attached to Atg5
in a manner similar to the ubiquitination. Although Atg12 has scarcely
sequence similarity with ubiquitin, its secondary structure was predicted
to have ubiquitin-like domain in the C-terminus region. We prepared
N-terminally truncated Atg12 mutant of the yeast S. cerevisiae according to the predicted secondary structure. Truncated form of
Atg12 having only a predicted ubiquitin-like domain conjugated with
Atg5 was still active in autophagy. While a truncated Atg12 mutant
lacking the first beta strand in the ubiquitin-like domain didn’t
conjugate with Atg5. These results showed that the ubiquitin-like
domain of Atg12 is necessary and sufficient for conjugation and autophagy.
Furthermore, we altered several hydrophobic amino acid residues in
the ubiqutin-like domain of Atg8 and found a certain amino acid residue
is critical for autophagy, even though still active for conjugation
with Atg5. In contrast to the ubiqutin/proteasome pathway, autophagy is thought
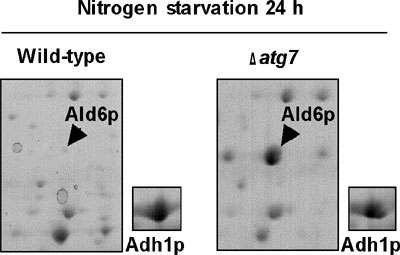
to be non-selective protein degradation process. We surveyed the changes
of protein profiles after nitrogen starvation of wild-type and Δatg7
cells using 2D-PAGE, and we found that cytosolic acetaldehyde dehydrogenase
(Ald6p; Figure 1) was degraded more rapidly than other cytosolic proteins
in an autophagy dependent manner. We observed that Ald6p was enclosed
in the autophagosome and delivered to the vacuole preferentially.
The Ald6p may be harmful to cells during nitrogen starvation, since
disruption of ALD6 improved the loss of viability of the Δatg7 mutant. This is the first report of the preferential
degradation of a detrimental protein via autophagy.
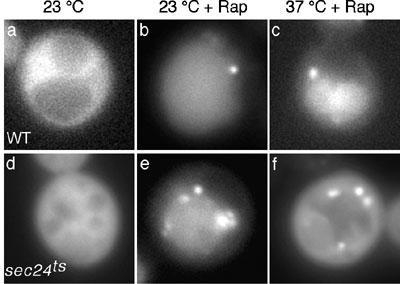
In addition to Atg proteins, we have previously identified the involvement
of secretory proteins in autophagy. Autophagosome formation is completely
blocked when some early Sec proteins such as Sec24p, are defective,
which are involved in the formation of the COPII coated vesicles from
ER to Golgi. The autophagic defect in sec24 deleted mutant
cells was, however, suppressed upon the recovery of ER-Golgi secretory
flow by the overexpression of its homologue, Sfb2p. We found that
the autophagic defect is not observed in sec13 and sec31
mutants, a phenomenon that can be explained by the fact that starvation
stress suppresses the secretory defect of these mutants. These observations
indicate that the active flow in the early secretory pathway plays
an important role in autophagy. Both autophagy and its closely related
cytoplasm to vacuole-targeting (Cvt) pathway occur through a pre-autophagosomal
structure (PAS), and since the PAS and the functional Cvt pathway
exist in all sec mutants, the early secretory pathway must be involved
specifically in autophagy, subsequent to PAS formation.
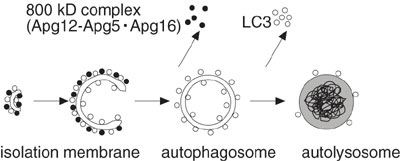
We have shown that, in yeast and mammalian cells, the Atg12-Atg5 protein
conjugate, which is formed by a ubiquitin-like system, is essential
for autophagosome formation. In yeast, the Atg12-Atg5 conjugate interacts
with a small coiled-coil protein, Atg16, to form a ~350-kDa multimeric
complex. We demonstrate that the mouse Atg12-Atg5 conjugate forms
a ~800-kDa protein complex containing a novel WD repeat protein (Fig.
3). As the N-terminal region of this novel protein shows homology
with yeast Atg16, we have designated it mouse Atg16-like protein (Atg16L).
Atg16L, however, has a large C-terminal domain containing seven WD
repeats, absent from yeast Atg16. Atg16L interacts with both Atg5
and additional Atg16L monomers; neither interaction, however, depends
on the WD-repeat domain. In conjunction with Atg12-Atg5, Atg16L associates
with the autophagic isolation membrane for the duration of autophagosome
formation. As these features are similar to yeast Atg16, we concluded
Atg16L is the functional counterpart of the yeast Atg16. We also found
that membrane targeting of Atg16L requires Atg5, but not Atg12. As
WD repeat proteins provide a platform for protein-protein interactions,
the ~800-kDa complex is expected to function in autophagosome formation,
further interacting with other proteins in mammalian cells.
In yeast, autophagy is required for cell survival during starvation
and is necessary for spore formation. In contrast, the role of autophagy
in mammals is still poorly understood. Although the possible involvement
of autophagy in development, cell death and pathogenesis has been
repeatedly pointed out, systematic analysis has not been performed,
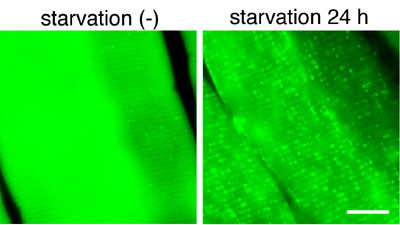
mainly due to a limitation of monitoring methods. Our recent studies
have made available several marker proteins for autophagosomes. To
understand where and when autophagy occurs in vivo, we have generated
transgenic mice systemically expressing GFP fused to LC3, which is
a mammalian homologue of yeast Aut7/Atg8. Cryosections of various
organs were prepared and the occurrence of autophagy was examined
by fluorescence microscopy (Fig. 4). Active autophagy was observed
in various tissues, such as the skeletal muscle, liver, heart, exocrine
glands, thymic epithelial cells, lens epithelial cells and podocytes.
Autophagy is differently induced by nutrient starvation in most tissues.
In some tissues, autophagy even occurs spontaneously. Our results
suggest that the regulation of autophagy is organ-dependent and the
role of autophagy is not restricted to the starvation response. This
transgenic mouse is a useful tool to study mammalian autophagy.
So far, autophagy in plant has been described by morphological studies.
Recent genome-wide search revealed significant conservation in autophagy
genes between yeast and plant. But little is known about the physiological
roles and molecular mechanisms underlying autophagy in higher plants.
To elucidate the plant autophagy, we focused ubiquitination-like
Atg8 lipidation system. In yeast, Atg8 binds to autophagosomes and
is delivered to the vacuole through autophagic process. Therefore,
Atg8 is a useful molecular marker for monitoring autophagic process.
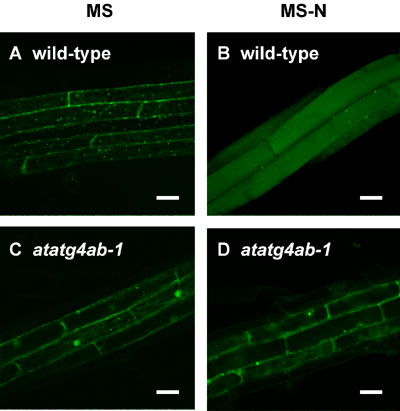
To establish a system monitoring autophagy in a whole plant, we
generated transgenic Arabidopsis expressing GFP-AtATG8 fusion protein.
In wild-type plants, GFP-AtATG8s were observed as ring structures
in the cytoplasm (Figure 5 panel A), and delivered to the lumens
of vacuole under nitrogen-starvation condition (Figure 5 panel B).
On the basis of analogy of yeast, we regarded these ring structures
as autophagosomes. In contrast, in double disruptant of AtATG4s
which are required for C-terminal cleavage of Atg8, GFP-AtATG8s
were not localized on autophagosomes (Figure 5 panel C) and were
not delivered to the vacuole under nitrogen-starvation condition
(Figure 5 panel D). These results indicate that AtATG8 is a suitable
marker for monitoring autophagy in plant.
|
||||||||||
Publication List: Mizushima, N., Kuma, A., Kobayashi, Y., Yamamoto, A., Takao, T., Natsume, T., Ohsumi, Y., and Yoshimori, T., Mouse Apg16L, a novel WD repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci., 116, 1679-1688 (2003) Hamasaki, M., Noda, T., and Ohsumi Y. The early secretory pathway contributes to autophagosome formation. Cell Struc. Func., 28, 49-54 (2003) Sugawara, K., Suzuki, N., Fujioka, Y., Mizushima, N., Ohsumi, Y., Inagaki, F. Crystalization and preliminary X-ray analyses of LC3-I, Acta Crys., D59, 1464-1465, (2003) Mukaiyama, H., Baba, M., Osumi, M., Aoyagi, S., Kato, N., Ohsumi, Y., Sakai, Y. Modification of a Ubiquitin-like protein Paz2 conducted micropexophagy through formation of a membrane structure. Mol. Biol. Cell., in press (2003) Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B., Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest., 112, 1809-1820 (2003) Klionsky DJ, Cregg JM, Dunn WA Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. Dev. Cell., 5, 539-45 (2003) Mizushima, N., Yamamoto, A., Matsui, M., Yoshimori, T. and Ohsumi, Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell, in press (2004) Ohashi, M., Mizushima, N., Kabeya, Y. and Yoshimori, T. Localization of mammalian NAD (P) H steroid dehydrogenase-like protein on lipid droplets J. Biol. Chem. 278, 36819-36829 (2003) Mizushima, N., Yamamoto, A., Matsui, M., Yoshimori, T. and Ohsumi, Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell in press Mizushima, N., Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. In press (2004) Ohsumi, Y. Cellular recycling system- molecular mechanism of autophagy. In Cell Growth, Cold Spring Harbor Press, in press (2003) Kamada, Y., Sekito,T., Ohsumi Y., Autophgy in yeast: a TOR-mediated response to nutrient starvation. Curr Top Microbiol Immunol., 279, 73-84 (2004) Ohsumi, Y. and Mizushima, N. Two ubiquitin-like conjugation systems essential for autophagy. Sem. Cell Dev. Biol., in press (2004) |