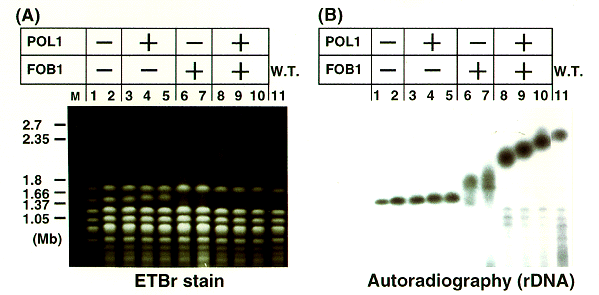
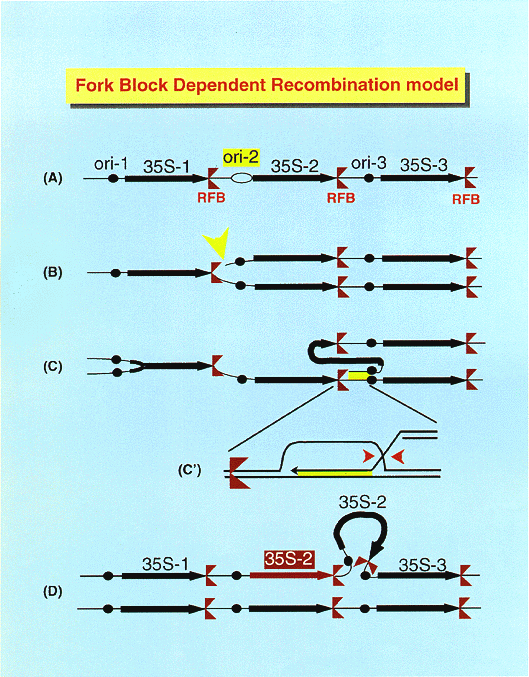
NATIONAL INSITUTE FOR BASIC BIOLOGY

National Institute for Basic Biology
DIVISION OF GENE EXPRESSION AND REGULATION II
- Professor:
- Takashi Horiuchi
- Research Associates:
- Masumi Hidaka
- Takehiko Kobayashi
- Ken-ichi Kodama
- Katsuki Johzuka
- Takehiko Kobayashi
- Post Doctoral Fellow:
- Katsufumi Ohsumi
- Keiko Taki
- Graduate Students:
- Hiroko Urawa
- Technical Staff:
- Kohji Hayashi
- Yasushi Takeuchi