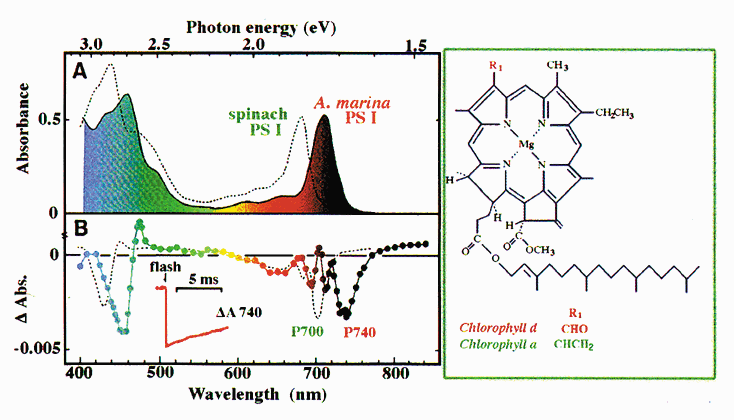
NATIONAL INSITUTE FOR BASIC BIOLOGY

National Institute for Basic Biology
OFFICE OF DIRECTOR
- Director-General:
- Hideo Mohri
- Associate Professors:
- Shigeru Itoh
- Ryuji Kodama
- Kohji Ueno
- Ryuji Kodama
- Research Associate:
- Akinao Nose (- May 31, 1998)
- Masuo Goto (on leave)