NATIONAL INSITUTE FOR BASIC BIOLOGY

National Institute for Basic Biology
DIVISION OF BIOLOGICAL REGULATION AND PHOTOBIOLOGY
(Adjunct)
- Associate Professor (adjunct):
- Hirokazu Kobayashi
- Research Associate:
- Noritoshi Inagaki
NATIONAL INSITUTE FOR BASIC BIOLOGY

National Institute for Basic Biology
|
|
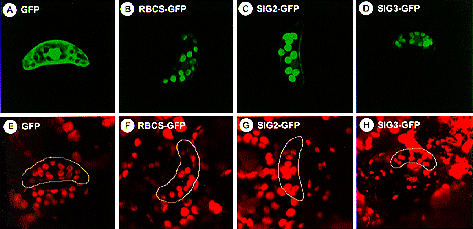
Fig. 1. Localization of GFPs fused to N-terminal regions of SIG2 and SIG3. GFP fusion constructs with the N-terminal regions of SIG2 ORF (SIG2-GFP, panels C and G) and SIG3 ORF (SIG3-GFP, panels D and H), and the transit peptide of small subunit of Rubisco (RBCS-GFP, panels B and F), as well as GFP alone (GFP, panels A and E), were introduced into tobacco leaves by particle bombardment. Guard cells were observed using MRC-1024 Confocal Imaging System (X 480) with excitation at 488 nm and emission at 520 nm (panels A-D), as well as excitation at 647 nm and emission at 666 nm (panels E-H). The same objects are shown in each pair of upper and lower panels. |




webmaster@nibb.ac.jp
Last Modified: 12:00, May 28, 1998