National Insitute for Basic Biology

National Insitute for Basic Biology

|
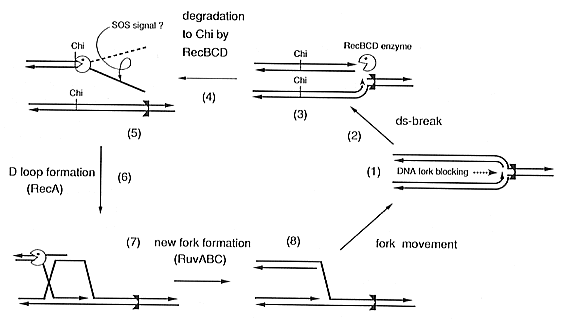
| Fig. 1.
Model of rescue from blocked DNA replication in E. coli, as applicable to a spontaneously stalled replication fork. Essentially the same sequential reactions would probably occur in S. cerevisiae. |